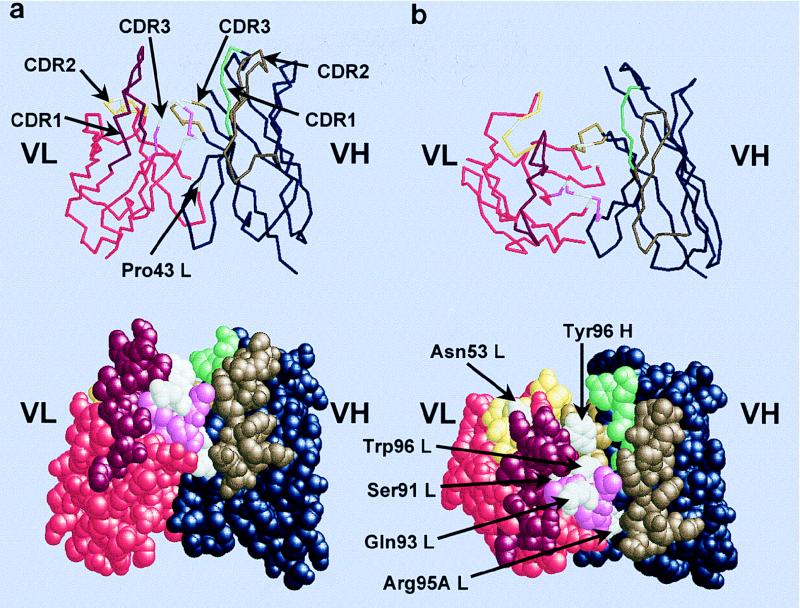
FIG. 8.
Model of the prototypic unmutated canonical HibCP antibody (Fv fragment). (a) Side view of the Fv fragment as a backbone and spacefill model. The CDRs form a groove in the antigen binding site because of the very short VH CDR3 and the long Vκ CDR1. The long linear HibCP molecule is expected to be placed in the groove. (b) Top view of the Fv fragment displaying the putative paratope for HibCP as well as the HibId-1 idiotope. The three-dimensional structure was predicted by using the ABGEN algorithm on the VH and VL amino acid sequences of Fab3-23gl/A2aJk1 (Fig. 1). Amino acids discussed in the text are indicated in white. The remaining residues in the VL sequence are indicated by the following colors: red, FRs; bordeaux, CDR1; yellow, CDR2; pink, CDR3. The remaining residues in the VH sequence are indicated by the following colors: blue, FRs; green, CDR1; brown, CDR2; curry, CDR3. Atomic coordinates are available from the Protein Data Bank (the Research Collaboratory for Structure Bioinformatics [RCSB]) under the entry 1HOU.