Abstract
Cryptosporidium parvum is a zoonotic parasite of the intestine and poses a threat to human and animal health. However, it is difficult to obtain a large number of oocysts for genome sequencing using in vitro culture. To address this challenge, we employed the strategy of whole-genome amplification of 10 oocysts followed by long-read sequencing and obtained a high-quality genome assembly of C. parvum IIdA19G1 subtype isolated from a pre-weaning calf with diarrhea. The assembled genome was 9.13 Mb long and encompassed eight chromosomes with six capped by telomeric sequences at one or both ends. In total, 3,915 protein-coding genes were predicted, exhibiting a high completeness with 98.2% single-copy BUSCO genes. To our current knowledge, this represents the first chromosome-level genome assembly of C. parvum achieved through the combined use of whole-genome amplification of 10 oocysts and long-read sequencing. This achievement not only advances our understanding of the genomic landscape of this zoonotic intestinal parasite, but also provides valuable resources for comparative genomics and evolutionary analyses within the Cryptosporidium clade.
Subject terms: Genome informatics, Parasite genomics
Background & Summary
Cryptosporidium spp. are parasitic apicomplexans that cause moderate-to-severe diarrhea in humans and animals1. The lack of widely efficacious medications and the absence of a vaccine necessitate heavy reliance on infection prevention for the management of cryptosporidiosis, thereby highlighting the urgent requirement for innovative interventions2,3. Cryptosporidium species have been detected in 155 mammalian species, including primates4,5. Currently, at least 44 species of Cryptosporidium have been identified6. Several species, including Cryptosporidium parvum, Cryptosporidium ubiquitum, and Cryptosporidium muris, exhibit wide host ranges, leading to zoonotic infections in conjunction with other Cryptosporidium spp7. Whole-genome sequencing (WGS) and comparative genomic analysis have been employed to elucidate the genetic underpinnings responsible for variations in host range among different species of Cryptosporidium, as well as the process of host adaptation within each species8–10. The use of WGS analysis has become more prevalent in the characterization of Cryptosporidium owing to the emergence of next-generation sequencing (NGS) technologies. A total of 15 species have been subjected to genome sequencing, encompassing C. parvum, Cryptosporidium hominis, C. ubiquitum, Cryptosporidium meleagridis, and others. The majority of the available genomic sequence data (19 sequences) pertain to the zoonotic C. parvum, yet only two of these sequences have been annotated11. The initial comprehensive genome assembly for C. parvum Iowa II was made accessible in 2004 using a random shotgun sequencing technique. This approach yielded a total of 9.1 Mb of DNA sequences distributed across all eight chromosomes12. According to previous studies, the genetic divergence between C. parvum and C. hominis was estimated to be approximately 3%-5% at the DNA level13.
One of the primary challenges encountered in genomics research on Cryptosporidium spp. is the limited availability of adequately purified oocysts in sufficient quantities for NGS analysis, primarily because of the absence of an in vitro culture system capable of propagating parasites. Previous WGS analyses of Cryptosporidium have been conducted using oocysts purified from laboratory animals that were infected12,14,15. Troell et al.16 sequenced the Cryptosporidium single-oocyst genome, followed by a comprehensive whole-genome analysis through comparison with de novo assembly of the reference population genome. This research represents a significant milestone as it establishes the feasibility of acquiring high-quality genomic data from single-celled eukaryotes, encompassing both extensive coverage and precise information16. However, previous research on Cryptosporidium only involved single-oocyst NGS of the genome without assembling it at the chromosomal level.
Here, our study aimed to address this limitation by generating a reference genome for C. parvum using long-read sequencing data from Oxford nanopore technology (ONT) and PacBio high fidelity (HiFi) sequencing platforms, along with error correction using short-read data. As a result, the assembled genome of C. parvum was 9.13 Mb in length and showed a high completion rate with 98.2% single-copy BUSCO genes. A total of 3,915 protein-coding genes were predicted, of which 3,666 genes (93.6%) were functionally annotated. This study is an attempt to complete the high-quality chromosome-level genome assembly of Cryptosporidium species using 10 oocysts amplification coupled with long-read sequencing, which might also be an effective strategy for genome sequencing projects of other difficult-to-collect or uncultivable pathogens.
Methods
Sample collection and genome sequencing
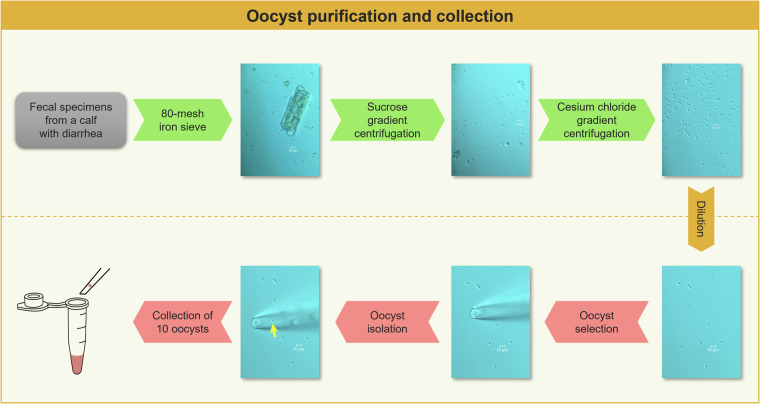
The Cryptosporidium strain was isolated from a calf with pre-weaning diarrhea in Henan, China, and identified as C. parvum using the SSU rRNA gene17. It was then subtyped by sequence analysis of the 60 kDa glycoprotein gene18 and identified as IIdA19G1 subtype. Oocysts of the identified Cryptosporidium species were purified using a three-step filtering (Fig. 1) comprising raw fecal filtration using 80-mesh iron sieve, sucrose gradient centrifugation, and cesium chloride gradient centrifugation19,20. Purified Cryptosporidium oocyst fluid (6 μL) was absorbed using a 10 μL pipette and dripped onto a glass petri dish. Under an inverted Olympus microscope at 60 × (OLYMPUS-BX53, Japan), a single oocyst of C. parvum was isolated using a three-axis hydraulic micromanipulator (World Precision Instruments Inc., USA). In this study, 10 oocysts were selected and pooled into a PCR tube containing 4 μL PBS buffer (Fig. 1).
Fig. 1.
The purification and collection process of oocyst. (Yellow arrow: C. parvum oocyst).
The 10 oocysts sample was then lysed and whole-genome amplified using the REPLI-g Single Cell Kit (based on multiple displacement amplification method; QIAGEN, Germany). The resulting whole-genome amplification (WGA) products were purified using Agencourt AMPure XP beads (BECKMAN, USA) to remove dNTP, primers, primer dimers, salt ions, and other impurities from the amplified products. According to NanoDrop One (Thermo Fisher Scientific, USA), the WGA product concentration in C. parvum was 762 ng/μL. Through Qubit 3.0 (Invitrogen, USA), the quantity of the WGA product was 30 μg, and the Nc/Qc (NanoDrop/Qubit) value was 1.2.
The high-quality amplified DNA was used to construct the genomic library, and the library was size-selected using BluePippin (Sage Science, USA). The purified and size-selected library was then sequenced on the Pacific Biosciences Sequel II platform (HiFi) in continuous long-read mode (Pacific Biosciences, USA) and the PromethION 48 sequencer (ONT, UK) following the manufacturer’s instructions, respectively. A total of 3.5 Gb (386 × coverage) PacBio HiFi and 8.8 Gb (967 × coverage) ONT long sequencing reads were obtained after removing adaptors and chimeric reads (Table 1). For short-read sequencing, library preparation was performed with 50 ng of fragmented DNA using the MGIEasy Universal DNA Library Prep Kit (MGI, Shenzhen, China) and then sequenced on the MGISEQ-2000 platform (BGI, Shenzhen, China). About 1.6 Gb (173 × coverage) of 150-bp paired-end reads (clean data) were generated using MGI sequencing platform (Table 1).
Table 1.
Sequencing data used for the genome assembly of C. parvum.
| Sequencing technology | MGI | PacBio | ONT |
|---|---|---|---|
| Clean data (Gb) | 1.6 | 3.5 | 8.8 |
| Reads Mean (bp) | 150 | 4,949 | 5,807 |
| Reads N50 (bp) | 150 | 5,105 | 6,535 |
| Reads Max (bp) | 150 | 25,327 | 92,140 |
| Depth (×) | 173 | 386 | 967 |
| GC content (%) | 32.2 | 31.1 | 31.9 |
De novo assembly
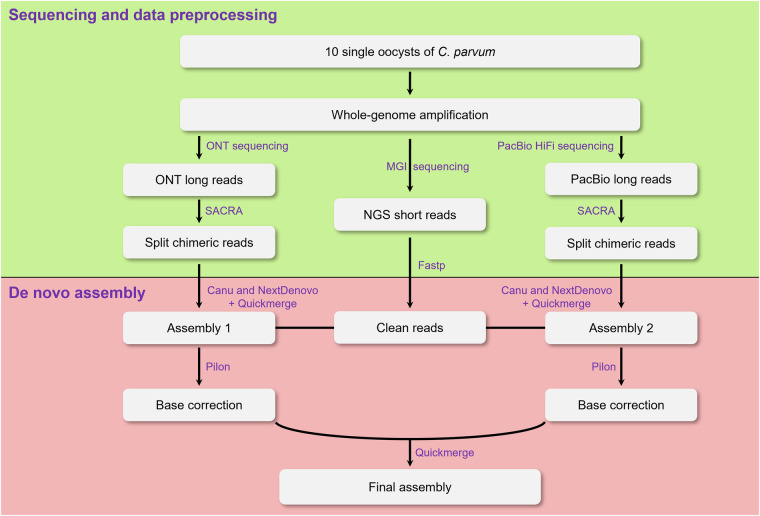
We first used SACRA v.2.021 to split chimeric long reads derived from multiple displacement amplification and fastp v.0.20.122 to trim adapter and low-quality bases in short reads. 486,818 chimera-containing reads in PacBio data and 1,394,568 in ONT data were identified and split using SACRA v.2.0, respectively. The clean long reads from ONT and PacBio platforms were independently assembled using Nextdenovo v.2.5.2 (https://github.com/Nextomics) and Canu v.2.2.223 with default parameters (Fig. 2). To improve the assembly contiguity, the outputs for each platform were merged using Quickmerge v.0.3 with default parameters (https://github.com/mahulchak/quickmerge). The merged assembly was then polished two rounds with Pilon v.1.24 (https://github.com/broadinstitute/pilon) using short clean reads24 (Fig. 2). For this, short reads were first mapped to the assembly using BWA v.0.7.1025 with default parameters. Then reads with mapping quality at least 30 were used for polishing (--minmq 30). The polished assemblies from the two sequencing platforms were further merged using Quickmerge v.0.3. Finally, we obtained a total genome length of 9.13 Mb across eight assembled contigs with six capped by telomeric repetitive sequences (TTTAGG)n at one or both ends (Table 2).
Fig. 2.
Framework of genome assembly.
Table 2.
Comparison between the assembled and published C. parvum reference genomes.
| Statistic | C. parvum (This study) | C. parvum (Iowa II68) | C. parvum (IOWA-ATCC69) |
|---|---|---|---|
| Number of contigs | 8 | 8 | 8 |
| Genome size (bp) | 9,128,570 | 9,102,324 | 9,122,263 |
| Largest contig (bp) | 1,336,160 | 1,344,712 | 1,332,634 |
| Contigs with two telomeres | 1 | 3 | 6 |
| Contigs with one telomere | 5 | 3 | 1 |
| N50 (bp) | 1,106,866 | 1,104,417 | 1,108,396 |
| GC (%) | 30.16 | 30.23 | 30.18 |
| Number of predicted genes | 3,915 | 3,886 | 4,424 |
| Complete BUSCOs (%) | 98.2 | 98.2 | 98.2 |
| Complete and single-copy BUSCOs (%) | 98.2 | 98.2 | 98.2 |
| Complete and duplicated BUSCOs (%) | 0.0 | 0.0 | 0.0 |
| Fragmented BUSCOs (%) | 0.4 | 0.4 | 0.6 |
| Missing BUSCOs (%) | 1.4 | 1.4 | 1.2 |
| Total Lineage BUSCOs | 502 | 502 | 502 |
The statistics of genome assembly, including contig length, N50 and GC content were comparable to those of the published C. parvum reference genome. Benchmarking Universal Single-Copy Orthologs (BUSCO) v.5.4.626 was used to evaluate the completeness of the C. parvum genome assembly against the Coccidia_odb10 database.
Gene prediction and annotation
Protein-coding genes were predicted through the integration of ab initio methods, homology alignment data, and transcriptomic data as described previously27. Briefly, the transcriptomic data28 for gene model training and protein data29 for homology alignment of C. parvum were downloaded from CryptoDB (https://cryptodb.org). For ab initio methods, PASA v.2.4.030 was applied to produce candidate gene structures, which could be applied to obtain a set of gene structures for training the SNAP (v.2013-11-29)31, Augustus v.3.3.332 (--genemodel=complete), GenomeThreader v.1.6.133, and GlimmerHMM v.3.0.434 using default parameters. Subsequently, Augustus v.3.3.332 and GlimmerHMM v.3.0.434 were used to predict gene structure using trained gene models. Gene models derived from ab initio and homologous alignment approaches was finally integrated into a non-repetitive gene set using EvidenceModeler v.1.1.135 and 3,915 protein-coding genes were predicted (Table 2).
The predicted protein sequences were functionally annotated through searching against 18 databases using InterProScan v.5.4536, including CDD37, Coils38, Gene Ontology39, Gene3D40, Hamap41, MobiDBLite42, PANTHER43, Pfam44, Phobius45, PIR46, PRINTS47, ProSite48, SFLD49, SignalP50, SMART51, SUPERFAMILY52, TIGRFAM53, TMHMM54 (Table 3). Finally, 3,666 genes (93.6% of the total) were successfully annotated.
Table 3.
Gene function annotation statistics of the assembled C. parvum genome.
| Database | Gene number | Percentage (%) |
|---|---|---|
| CDD | 1,027 | 26.2 |
| Coils | 1,076 | 27.5 |
| Gene Ontology | 1,963 | 50.1 |
| Gene3D | 2,161 | 55.2 |
| Hamap | 125 | 3.2 |
| MobiDBLite | 1,449 | 37.0 |
| PANTHER | 2,286 | 58.4 |
| Pfam | 2,299 | 58.7 |
| Phobius | 1,376 | 35.2 |
| PIR | 519 | 13.3 |
| PRINTS | 350 | 8.9 |
| ProSite | 1,687 | 43.1 |
| SFLD | 19 | 0.5 |
| SignalP | 577 | 14.7 |
| SMART | 1,050 | 26.8 |
| SUPERFAMILY | 2,039 | 52.1 |
| TIGRFAM | 216 | 5.5 |
| TMHMM | 854 | 21.8 |
| All Annotated | 3,666 | 93.6 |
Noncoding RNAs annotation
Non-coding RNAs are usually divided into several groups, including rRNA, tRNA, miRNA, and snRNA. Identification of the rRNA genes was conducted by Barrnap v.0.955 using default parameters. The tRNAscan-SE v.2.0.1256 was used to predict tRNA with eukaryote parameters. The miRNA genes were identified by searching miRBase v.21 databases57 using default parameters. The snRNA genes were predicted using INFERNAL v.1.158 based on Rfam v.12.0 database59 using default parameters. Finally, a total of 14 rRNAs, 45 tRNAs, 0 miRNA and 8 snRNAs were predicted (Table 4).
Table 4.
Noncoding RNA of the assembled genome.
| RNA classification | Number |
|---|---|
| rRNA | 14 |
| tRNA | 45 |
| miRNA | 0 |
| snRNA | 8 |
Data Records
The raw sequencing data, including MGI short reads (accession CRA01331560), PacBio HiFi (accession CRA01331661) and ONT long reads (accession CRA01332062), and the whole-genome assembly (accession GWHEQBI0000000063) of the C. parvum IIdA19G1 strain can be access through National Genomics Data Center, China National Centre for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (PRJCA02054064). The genome assembly65 have also been submitted to NCBI database under the BioProject accession number PRJNA1045063. Moreover, the genomic annotation results have been deposited in the Figshare database66.
Technical Validation
We evaluated the assembly using two criteria: the mapping of short and long sequencing reads and BUSCO assessment. The reads from the short-insert library were re-mapped onto the assembly using BWA v.0.7.1025, while PacBio HiFi and ONT long reads were aligned using minimap2 v.2.2467 using default parameters. The assembly completeness was evaluated using BUSCO v.5.4.626 using the Coccidia dataset and genome mode (-l coccidia_odb10 -m geno).The mapping rate for short reads was 99.4%, while the mapping rates for HiFi and ONT long reads were 99.6% and 97.7%, respectively (Table 5). Moreover, 98.2% of the complete single-copy BUSCO genes were included in the assembled genome (Table 2). Overall, these assessments independently confirmed the accuracy and completeness of the genome assembly.
Table 5.
Results of long and short sequencing reads mapped to the assembled C. parvum genome.
| Sequencing platform | MGI | PacBio | ONT |
|---|---|---|---|
| Total reads (bp) | 1,576,020,900 | 3,519,749,056 | 8,825,522,949 |
| Mapped reads (bp) | 1,566,605,400 | 3,505,499,876 | 8,622,067,393 |
| Mapping rate (%) | 99.4 | 99.6 | 97.7 |
Acknowledgements
This research was funded by the National Key Research and Development Plan Project (2022YFD1800200), NSFC-Henan Joint Fund Key Project (U1904203), and Leading Talents of the Central Plains Thousand Talents Program (19CZ0122). We thank the members of Protist 10,000 Genomes Project (P10K) consortium for their helpful suggestions. The bioinformatics analysis was supported by the Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences, China. We also thank the LetPub Editor for editing the language of this manuscript.
Author contributions
Conceived and Designed: L.X.Z. and G.Y.W. Manuscript: Y.C.C. and L.X.Z. Analysis: Y.C.C., J.Y.H., G.Y.W., K.C. and J.X. Reagents/materials: H.K.Q., K.H.Z., Y.F., J.Q.L. and R.J.W. Supervision: J.Y.H., W.M., G.Y.W., and L.X.Z. All of the authors have read and approved the final manuscript.
Code availability
No custom code was used in this study. The data analyses used standard bioinformatic tools specified in the methods.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guangying Wang, Email: wangguangying@ihb.ac.cn.
Longxian Zhang, Email: zhanglx8999@henau.edu.cn.
References
- 1.Kotloff, K. L. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet382, 209–222 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Bhalchandra, S., Cardenas, D. & Ward, H. D. Recent Breakthroughs and Ongoing Limitations in Cryptosporidium Research. F1000 Res.7, F1000 Faculty Rev-1380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavez, M. A. & White, A. C. J. Novel treatment strategies and drugs in development for cryptosporidiosis. Expert. Rev. Anti. Infect. Ther.16, 655–661 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Fayer, R., Morgan, U. & Upton, S. J. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol.30, 1305–1322 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Fayer, R. Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol.126, 37–56 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Ryan, U. M. et al. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia - a 50 year perspective (1971-2021). Int. J. Parasitol.51, 1099–1119 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Ryan, U., Zahedi, A. & Paparini, A. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol.38, 535–547 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Fan, Y. Y., Feng, Y. Y. & Xiao, L. H. Comparative genomics: how has it advanced our knowledge of cryptosporidiosis epidemiology? Parasitol. Res.118, 3195–3204 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Khan, A., Shaik, J. S. & Grigg, M. E. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop.184, 1–14 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Kim, K. U. et al. Comparison of functional gene annotation of Toxascaris leonina and Toxocara canis using CLC genomics workbench. Korean. J. Parasitol.51, 525–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baptista, R. P. et al. Long-read assembly and comparative evidence-based reanalysis of Cryptosporidium genome sequences reveal expanded transporter repertoire and duplication of entire chromosome ends including subtelomeric regions. Genome. Res.32, 203–213 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahamsen, M. S. et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science304, 441–445 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Mazurie, A. J. et al. Comparative genomics of Cryptosporidium. Int. J. Genomics2013, 832756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widmer, G. et al. Comparative genome analysis of two Cryptosporidium parvum isolates with different host range. Infect. Genet. Evol.12, 1213–1221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu, P. et al. The genome of Cryptosporidium hominis. Nature431, 1107–1112 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Troell, K. et al. Cryptosporidium as a testbed for single cell genome characterization of unicellular eukaryotes. Bmc Genomics17, 471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao, L. H. et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol.65, 3386–91 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alves, M. et al. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol.41, 2744–2747 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heyman, M. B., Shigekuni, L. K. & Ammann, A. J. Separation of Cryptosporidium oocysts from fecal debris by density gradient centrifugation and glass bead columns. J. Clin. Microbiol.23, 789–791 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilani, R. T. & Sekla, L. Purification of Cryptosporidium oocysts and sporozoites by cesium chloride and Percoll gradients. Am. J. Trop. Med. Hyg.36, 505–508 (1987). [DOI] [PubMed] [Google Scholar]
- 21.Kiguchi, Y. et al. Long-read metagenomics of multiple displacement amplified DNA of low-biomass human gut phageomes by SACRA pre-processing chimeric reads. DNA Res.28, dsab019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, S. et al. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res.27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, G. et al. A strategy for complete telomere-to-telomere assembly of ciliate macronuclear genome using ultra-high coverage Nanopore data. Comput. Struct. Biotechnol. J.19, 1928–1932 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simao, F. A. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Jiang, C. et al. iGDP: An integrated genome decontamination pipeline for wild ciliated microeukaryotes. Mol. Ecol. Resour.23, 1182–1193 (2023). [DOI] [PubMed] [Google Scholar]
- 28.CryptoDB sequence read archive.https://cryptodb.org/common/downloads/release-46/CparvumIowaII/fasta/data/CryptoDB-46_CparvumIowaII_AnnotatedTranscripts.fasta (2019).
- 29.CryptoDB sequence read archivehttps://cryptodb.org/common/downloads/release-46/CparvumIowaII/fasta/data/CryptoDB-46_CparvumIowaII_AnnotatedProteins.fasta (2019).
- 30.Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korf, I. Gene finding in novel genomes. BMC Bioinformatics5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mario, S. & Burkhard, M. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res.33, W465–467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gremme, G. et al. Engineering a software tool for gene structure prediction in higher organisms. Inform. Software Tech.47, 965–978 (2005). [Google Scholar]
- 34.Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Bio.9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics.30, 1236–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler-Bauer, A. et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res.41, D348–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitzkee, N. C., Fleming, P. J. & Rose, G. D. The Protein Coil Library: a structural database of nonhelix, nonstrand fragments derived from the PDB. Proteins58, 852–4 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet.25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeats, C. et al. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res.36, D414–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima, T. et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res.37, D471–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Necci, M. et al. MobiDB-lite 3.0: fast consensus annotation of intrinsic disorder flavors in proteins. Bioinformatics36, 5533–5534 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Mi, H. et al. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res.44, D336–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res.42, D222–D230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Käll, L., Krogh, A. & Sonnhammer, E. L. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res.35, W429–32 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barker, W. C. et al. The PIR-International Protein Sequence Database. Nucleic Acids Res.27, 39–43 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Attwood, T. K. et al. The PRINTS database: a fine-grained protein sequence annotation and analysis resource–its status in 2012. Database (Oxford)2012, bas019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sigrist, C. J. et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res.38, D161–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akiva, E. et al. The Structure-Function Linkage Database. Nucleic Acids Res.42, D521–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teufel, F. et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol.40, 1023–1025 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res.46, D493–D496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, D. et al. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res.35, D308–13 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haft, D. H., Selengut, J. D. & White, O. The TIGRFAMs database of protein families. Nucleic Acids Res.31, 371–3 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krogh, A. et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol.305, 567–80 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Loman, T. A Novel Method for Predicting Ribosomal RNA Genes in Prokaryotic Genomes., (2017).
- 56.Lowe, T. M. & Eddy, S. R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res.25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grifths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A. & Enright, A. J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res.34, D140–D144 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grifths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res.33, D121–D124 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.NGDC/CNCB Genome Sequence Archivehttps://ngdc.cncb.ac.cn/gsa/browse/CRA013315 (2024).
- 61.NGDC/CNCB Genome Sequence Archivehttps://ngdc.cncb.ac.cn/gsa/browse/CRA013316 (2024).
- 62.NGDC/CNCB Genome Sequence Archivehttps://ngdc.cncb.ac.cn/gsa/browse/CRA013320 (2024).
- 63.NGDC/CNCBhttps://ngdc.cncb.ac.cn/gwh/Assembly/82943/show (2023).
- 64.NGDC/CNCBhttps://ngdc.cncb.ac.cn/bioproject/browse/PRJCA020540 (2024).
- 65.NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_040285075.1 (2023).
- 66.Chen, Y. C. et al. Genome annotation data for the Cryptosporidium parvum IIdA19G1 subtype, figshare. Dataset, 10.6084/m9.figshare.26088349.v3 (2024).
- 67.Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.CryptoDB sequence read archive.https://cryptodb.org/common/downloads/release-46/CparvumIowaII/fasta/data/CryptoDB-46_CparvumIowaII_Genome.fasta (2019).
- 69.CryptoDB sequence read archive.https://cryptodb.org/common/downloads/release-46/CparvumIOWA-ATCC/fasta/data/CryptoDB-46_CparvumIOWA-ATCC_Genome.fasta (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Chen, Y. C. et al. Genome annotation data for the Cryptosporidium parvum IIdA19G1 subtype, figshare. Dataset, 10.6084/m9.figshare.26088349.v3 (2024).
Data Availability Statement
No custom code was used in this study. The data analyses used standard bioinformatic tools specified in the methods.