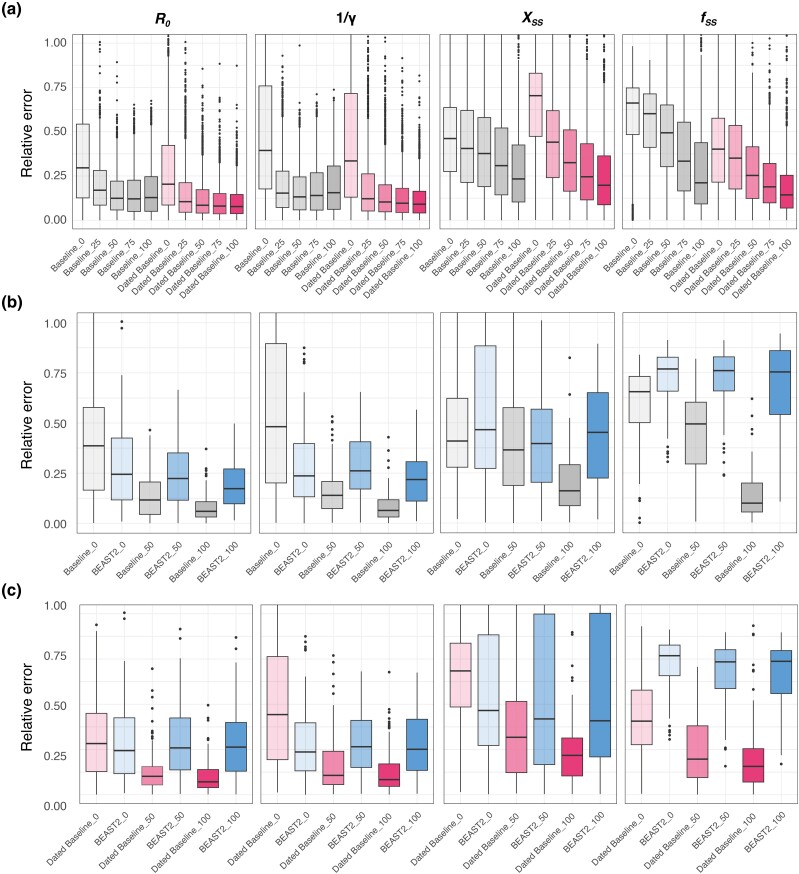
Fig. 4.
Performance comparison by incorporating varying levels of contact tracing data based on Baseline-Model, Dated Baseline-Model, and BEAST2. a) Comparison between the Baseline-Model and Dated Baseline-Model with varying levels of contact tracing data based on 1,000 simulated trees. The models are represented by Baseline-Model and Dated Baseline-Model, with the color intensity within each bar signaling the degree of contact tracing data integrated into the input trees. Darker shades denote a higher percentage of data incorporation. The term “Baseline_50” refers to the performance of the Baseline-Model with Genetic Polytomous Trees refined using 50% contact tracing data, encompassing cluster information and infection times. “Dated Baseline_50” indicates the performance of the Dated Baseline-Model with Genetic Polytomous Trees refined using 50% contact tracing data, including only cluster information. It is notable that the input trees are refined by infection time, with a 1-day time constraint margin using LSD2 (To et al. 2016), and an additional refinement with a stricter margin of 0.1 day, as shown in supplementary table S3, Supplementary Material online. b) Comparison between the Baseline-Model and BEAST2 (blue bar) with varying levels of contact tracing data (cluster information and infection times) based on 100 simulated trees. c) Comparison between Dated Baseline-Model and BEAST2 (blue bar) with varying levels of contact tracing data (cluster information only) based on 100 simulated trees. “BEAST2_50” indicates the performance of BEAST2 with Genetic Polytomous Trees refined using 50% contact tracing data, incorporating both cluster information and infection times in b), and only cluster information in c).