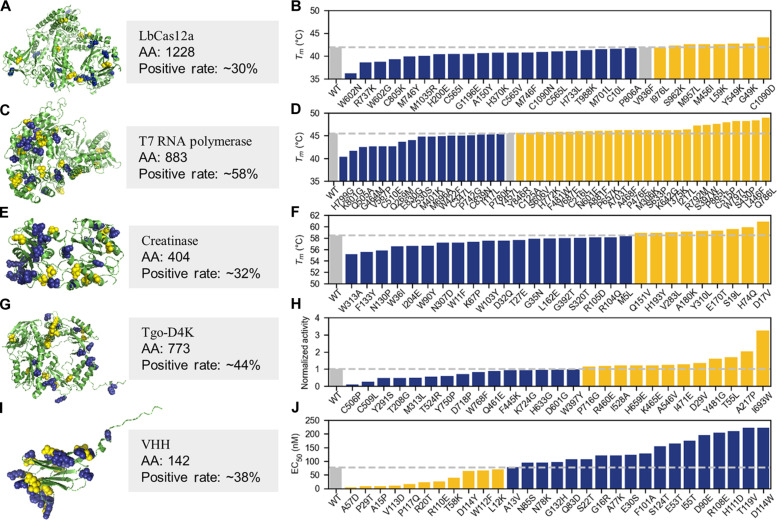
Fig. 3. Overview of the structures and performance results of single-site mutants predicted by the PRIME model.
The structures and experimental results of single-site mutants predicted by PRIME for LbCas12a (A and B), T7 RNA polymerase (C and D), creatinase (E and F), nonnatural nucleic acid polymerase (G and H) and VHH (I and J) are depicted. The data points representing the mutations were systematically arranged in ascending order, with the corresponding value for the wild-type protein delineated by a gray bar for comparative purposes. Mutants that exhibited superior performance compared to their wild-type counterparts in terms of targeted attributes are highlighted in yellow, while negative mutants are shown in blue. The engineering goals varied between proteins for practical purposes: for LbCas12a, T7 RNA polymerase, and creatinase, the objective was enhanced thermostability (Tm); for nonnatural nucleic acid polymerase (Tgo-D4K), the aim was to accelerate the synthesis rate of FANA; and for VHH, the goal was to improve the tolerance ability under extreme alkaline pH conditions [median effective concentration (EC50) of VHH binding to the antigen]. All mutated structure were folded by Alphafold2. Detailed experimental data can be found in the separate Excel file in the Supplementary Materials.