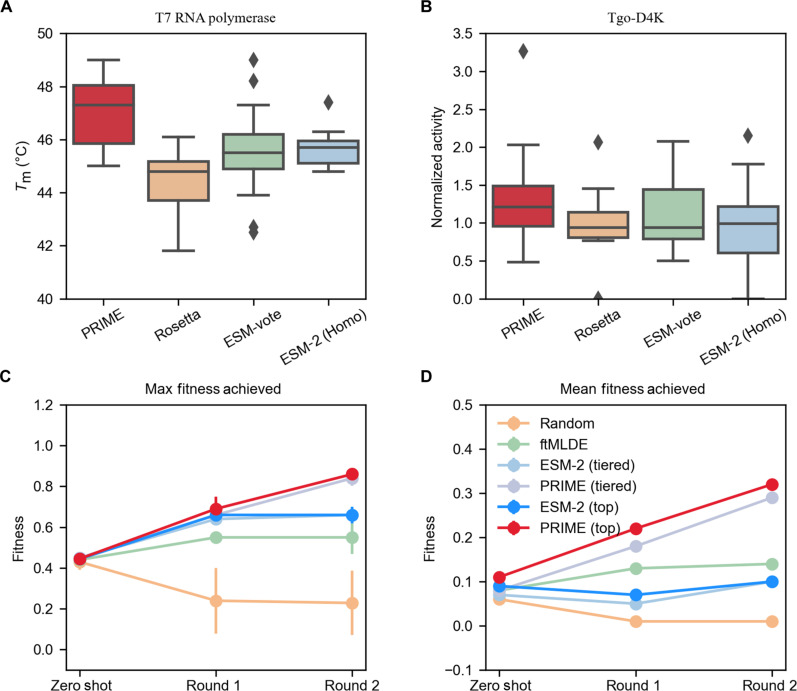
Fig. 4. Comparative analysis of PRIME and different models through wet-lab experiments and in silico benchmarking.
(A) Comparative web-lab results for the top 15 single-point mutations in T7 RNA polymerase. (B) Results for Tgo-D4K, as determined by PRIME, Rosetta, ESM-vote, and ESM2(homo). (C and D) Maximum (C) and mean (D) fitness outcomes obtained from in silico–directed evolution on the GB1 dataset, involving random mutagenesis, ftMLDE, ESM-2, and PRIME. For ESM-2 and PRIME, we examined both top-K sampling and the tiered sampling used by ftMLDE (77).