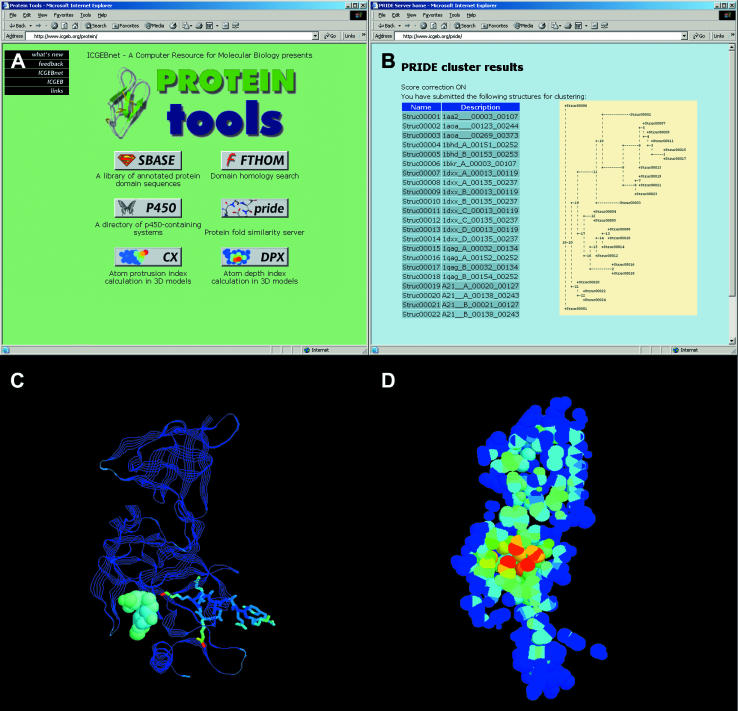
Figure 1.
The Protein tools server. (A) Title page [The SBASE (15), FTHOM (16) and P450 (17) services have been described elsewhere]. (B) Clustering of 24 CH domains by the PRIDE server. The tree is an ASCII rendering of the Newick file produced by the server. Bottom: structure of the human histone lysine N-methyltransferase SET7/9 complexed with a histone peptide and S-adenosyl-l-homocysteine (SAH), PDB: 1O9S. (C) Output of the CX server rendered with Rasmol (5). The enzyme is shown as ribbons, the peptide as sticks and SAH as a CPK model using; the structure is colored according to the cx values, calculated using a sphere radius of 8 Å. (D) Output of the DPX server rendered with Rasmol (5), The CPK model of the enzyme is shown in slab mode in the same orientation as in the left panel, and atoms colored according to their dpx values.