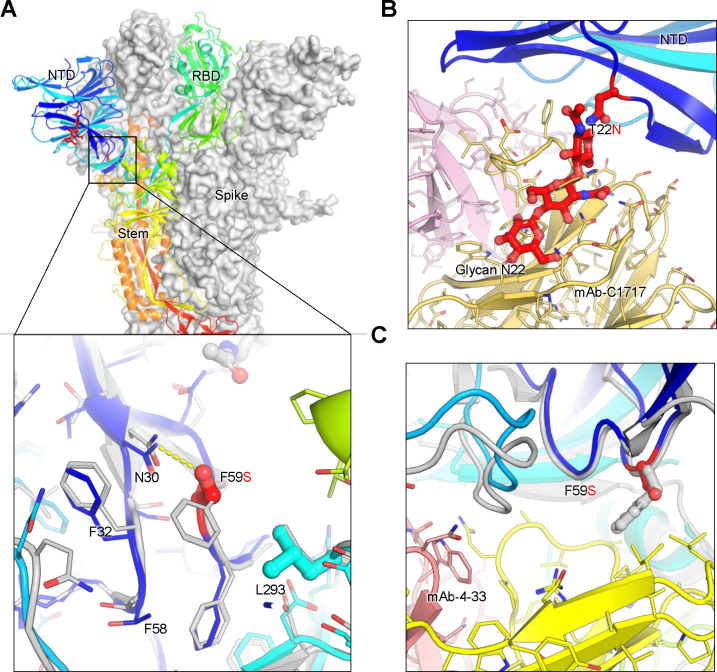
Figure 6. Structural modeling of key mutations in XEC spike.
(A) Structural representation of the spike protein domains, with the location of NTD mutations T22N and F59S highlighted. The spike is shown with two protomers displayed as a grey surface and one as a ribbon in rainbow colors. Inset: The F59S mutation alters its side-chain interaction with several nearby hydrophobic residues, including F32, F58, and L293, while introducing a hydrogen bond with residue N30. (B) The glycosylation at N22 (shown as sticks) interferes with the recognition of certain NTD-targeting antibodies, such as C1717, potentially reducing antibody binding efficiency. (C) The F59S mutation disrupts the epitopes of NTD-targeting antibodies, such as 4–33, by abolishing the interaction with a hydrophobic cluster, thereby impairing antibody recognition and contributing to immune evasion.