Figure 1: Blackbird outline.
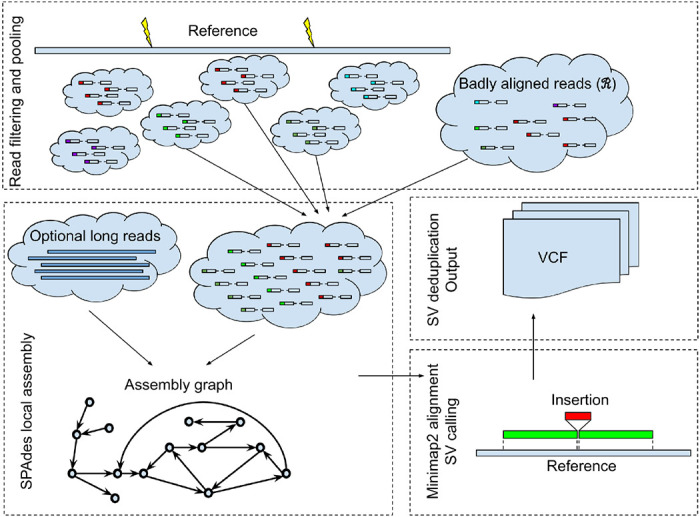
The reference genome is split into segments. Inside each segment, we select a set of barcodes and collect reads with these barcodes both aligned into the segment and unaligned. Different barcodes are shown by different colors attached to reads. Then we perform a local assembly for the segment and add long-reads aligned to the segment if they are available. The assembled contigs are mapped back to the reference genome segment using Minimap2, and variants are called from the alignments’ CIGAR strings. At the final step, calls are sorted, deduplicated, and stored in the VCF format.
