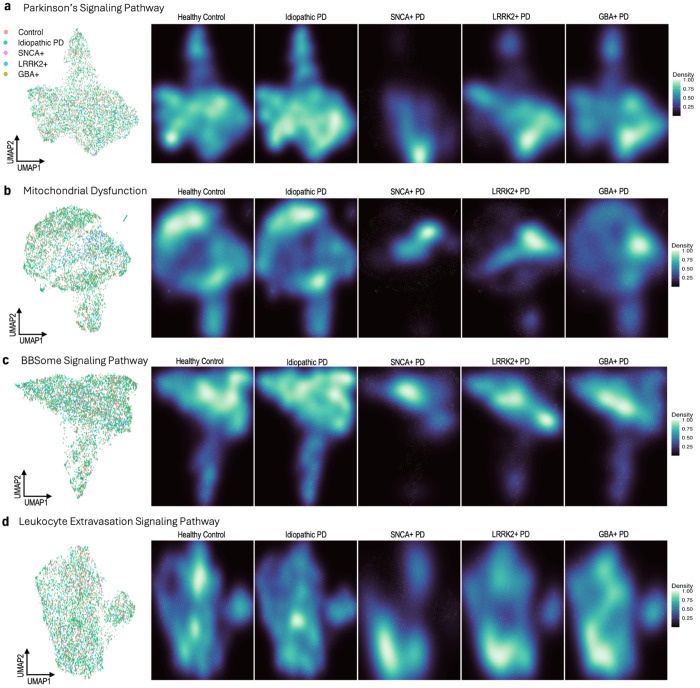
Fig. 5 |. UMAP dimensionality reduction with pathway-specific genes by disease status and genetic cohort.
UMAP embeddings were created from variant stabilizing transformed (VST) gene counts. Counts were then corrected for participant age and sex, as well as sample mRNA percentage and predicted neutrophil percentage. a, UMAP of 36 genes found in the Parkinson’s Signaling Pathway, with samples labeled by disease and genetic status, excluding samples from participants with unknown genetic status (n = 5,470). Corresponding density plots were made, stratified by disease and genetic status. b, A UMAP and set of density plots were created from 143 genes in the Mitochondrial Dysfunction pathway (n = 5,339). A small cluster of samples, mostly Control and Idiopathic PD, were removed from the Mitochondrial Dysfunction UMAP for visualization purposes. c, A UMAP of 59 BBSome Signaling Pathway genes and density plots (n = 5,358). A small cluster of samples, mostly Control and Idiopathic PD, were removed for visualization purposes. d, UMAP and density plots of 52 Leukocyte Extravasation Signaling Pathway genes (n = 5,345). A small cluster of samples, mostly Control and Idiopathic PD, were removed for visualization purposes.