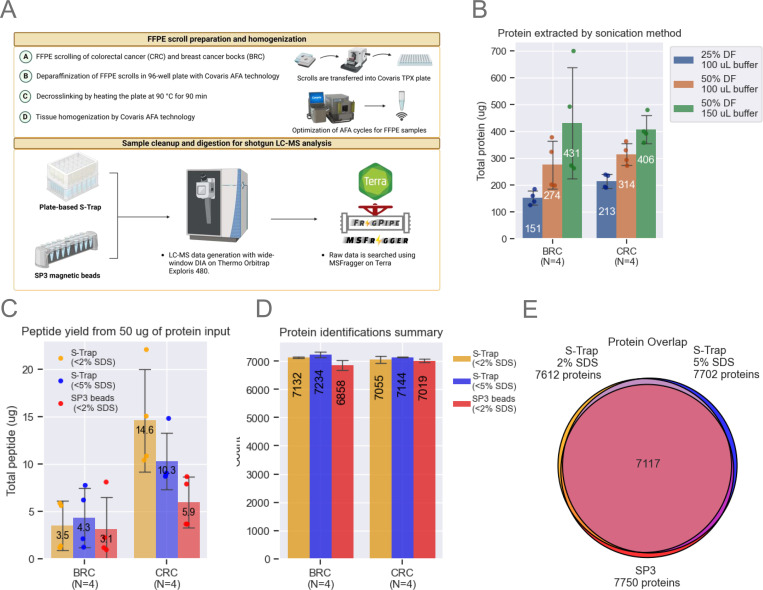
Figure 1: Method Optimization.
A. The FFPE workflow processes entire scrolls using a Covaris TPX plate. Breast cancer (BRC) and colorectal cancer (CRC) blocks were scrolled and transferred into a Covaris plate containing tissue lysis buffer. Sonication conditions were optimized to maximize protein extraction. SP3 beads and S-Trap plates were evaluated, starting with 50 μg of protein. Samples were analyzed on a Neo Vanquish LC coupled to an Exploris 480 (Thermo).
B. Optimization of protein extraction efficiency by adjusting sonication duty factors (DF) and buffer volume. Bar plots represent data from n=4 scrolls per block, showing means and standard deviation (SD) error bars.
C. Comparison of SP3 and S-Trap methods for digestion and cleanup using a fluorometric peptide assay. Bar plots represent data from n=4 scrolls per block, with means and SD error bars.
D. Unique protein identifications using MSFragger-DIA across the three tested workflows. Bar plots represent data from n=4 scrolls per block, with means and SD error bars.
E. Venn diagram illustrating protein overlap, with each circle representing the total number of unique proteins identified by each tested workflow.