Abstract
The extracellular matrix (ECM) is a mixture of glycoproteins and fibrous proteins that provide the biophysical properties necessary to maintain cellular homeostasis. ECM integrity is of particular importance during development, where it allows proper migration and cellular differentiation. Laminins are ECM heterotrimeric proteins consisting of α, β, and γ chains. There are five known α chains, four β chains, and three γ chains. Thus, there are 60 potential combinations for laminin trimers, however only 16 laminin trimers have been identified to date. Furthermore, none of them contain laminin β4 and its function is unknown. Here, we sought to characterize the role of LAMB4 (the gene encoding laminin β4) during human embryonic development of the peripheral sensory nervous system. Using human pluripotent stem cells (hPSCs), we found that LAMB4 is expressed in the ectoderm in the early stages of sensory neuron (SN) specification. SNs, part of the peripheral nervous system, are specialized neurons that detect pain, temperature, and touch. Surprisingly, more than 20 million people in the US have some form of peripheral nerve damage (including SNs), however there are very few treatment options available. Learning about the biology of peripheral neurons will uncover potential new therapeutic targets, thus we focused on understanding the effects of LAMB4 in SNs. First, we knocked out LAMB4 in hPSCs, using CRISPR/Cas9, and found that loss of LAMB4 impairs the migration of the SN progenitors neural crest cells (NCCs) and harms SN development and survival. To assess if LAMB4 has clinical relevance, we studied the genetic disorder Familial Dysautonomia (FD), which specifically affects the peripheral nervous system. FD is caused by a mutation in ELP1 (a component of the Elongator complex) leading to developmental and degenerative defects in SNs. A previous report showed that patients with severe FD harbor additional single nucleotide variants in LAMB4. We found that these variants sharply downregulate the expression of LAMB4 and laminin β4 levels in SNs differentiated from induced pluripotent stem cells (iPSCs) reprogramed from patients with severe FD. Moreover, a healthy ECM is sufficient to rescue the developmental phenotypes of FD, further confirming that ECM defects contribute significantly to the etiology of FD. Finally, we found that LAMB4/laminin β4 is necessary for actin filament accumulation and it interacts with laminin α4 and laminin γ3, forming the laminin-443, a previously unreported laminin trimer. Together, these results show that LAMB4 is a critical, but largely unknown gene required for SN development and survival.
Introduction
The extracellular matrix (ECM) is a dynamic network of proteins, glycoproteins, and proteoglycans that act as a cellular scaffold, providing the ideal environment to promote and maintain cellular homeostasis1,2. The ECM is a very dynamic structure, and it is constantly remodeled to support cell homeostasis3 and provides biophysical and biochemical cues required for cellular migration, differentiation, and survival during development4. For instance, neural crest cells (NCCs) arise from the border of the neural plate and the non-neural ectoderm of the developing embryo5,6. NCCs then migrate to different regions of the embryo in a process regulated by ECM remodeling and gradient of morphogens7, giving rise to sensory neurons (SNs) and autonomic neurons (part of the peripheral nervous system), glial cells, endocrine cells, craniofacial cartilage and bone, pigment cells, among others8. Changes in the biophysical properties of the ECM affect NCC differentiation9.
Laminins are major components of the ECM10. They are heterotrimeric proteins consisting of α, β, and γ chains that are expressed in different developmental stages for specific functions. For example, laminin-511 and laminin-111 are the most abundant laminins present during early development11,12, whereas laminin-523 is expressed only in the retinal outer membrane13. Laminins interact with the transmembrane proteins integrins14. They are heterodimer receptors that connect the ECM to intracellular components, resulting in activation of signaling pathways and reorganization of the cellular cytoskeleton15.
There are five α chains, four β chains, and three γ chains, which can assemble up to 60 different trimers, however, only 16 have been identified15. Of those, laminin β4 (expressed by the gene LAMB4) is understudied and no laminin trimer containing the laminin β4 chain has been identified. Laminin β4 downregulation has been linked to diverticulitis, a disease of the peripheral nervous system16. Additionally, single nucleotide variants of LAMB4 have been identified in patients with severe symptoms of the genetic disease Familial dysautonomia (FD), which specifically affects the peripheral neurons. Thus, we hypothesized that LAMB4/laminin β4 expression is necessary for development and homeostasis of the peripheral nervous system and their progenitors, the NCCs.
To address this, we use human pluripotent stem cell (hPSC) technology, which allows us to study human development, including the study of cellular and molecular mechanisms in cells from all three germ layers endoderm, mesoderm, and ectoderm17,18. Additionally, cells obtained from patients can be reprogrammed into induced pluripotent stem cells (iPSCs), which contain the same genetic background as the originating patients and thus are invaluable for disease modeling19.
Here, we used hPSC technology and identified that LAMB4 is expressed in the peripheral nervous system, and it is necessary for NCC migration, and development and survival of SNs. Patients with severe symptoms of the peripheral neuropathy FD harbor mutations in the LAMB4 gene. We show that SNs differentiating from FD iPSCs express low levels of laminin β4. Additionally, we report that the ECM that is deposited by healthy cells is sufficient to rescue the developmental phenotypes observed in FD. Finally, we show that laminin β4 forms the laminin-443 and it is required for accumulation of actin filaments (F-actin) in SNs. Together, our results confirm that the ECM is critical for the development and function of SNs and that laminin β4 is required for SN development.
Results
Laminin β4 is expressed in early stages of sensory neuron differentiation
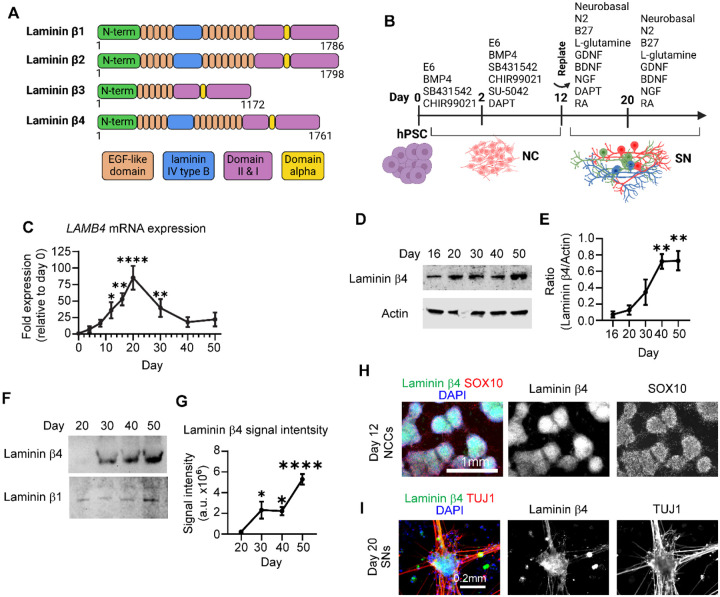
To understand the biological function of laminin β4 we sought to first assess the similarity between laminin β4 and the other laminin β chains. We started by asking whether metazoans express laminin β4. To do this, we compared the amino acid sequences of all the laminin β chains expressed in multiple species and generated a phylogenetic tree (Fig. S1A). We found that rodents (M. musculus and R. norvegicus) do not have a LAMB4 ortholog, whereas other vertebrates do, such as frogs (X. tropicalis), zebrafish (D. rerio), dogs (C. lupus), and chickens (G. gallus, Fig. S1A). Our results also showed that laminin β4 is closely related to laminin β1 and β2 (Fig. S1A), and LAMB4 is the ortholog expressed in the lowest number of the analyzed species: seven, compared to 11 for the other β chains. The lack of LAMB4 in rodents, particularly in mice, could be one of the reasons why LAMB4 has not been characterized yet and hints at its difficulty to study in vivo. We next asked whether laminin β4 shares similarities with other laminin β chains in humans. We didn’t find major differences between laminin β1, β2, and β4 chains, since they all shared the sequence of the N-terminal domain and 13 EGF-like domains (Fig. 1A and S1B). These domains are important for biological functions such as laminin network assembly, thus suggesting that they could bind similar proteins. In contrast, laminin β3 showed the shortest amino acid sequence and we only identified six EGF-like domains. On the C-terminal region, although all four laminin β chains showed similar length and number of domains, there were clear differences in the amino acid sequences (Fig. 1A and S1C). The C-terminal region bind to the α and γ chains, thus the differences between β chains might provide specificity during laminin assembly and be involved in the different affinities observed between laminin chains20. We next sought to understand which cell lineages express LAMB4. We first differentiated control hPSC-ctr-H9 cells into definitive endoderm (Fig. S2A). Although endoderm markers such as SOX17, GATA4, GATA6, and FOXA2 were highly expressed, LAMB4 was not (Fig. S2B). We also analyzed RNAseq of hindgut differentiated from hPSCs21, where we found that LAMA1, LAMA5, LAMB1, LAMB2, LAMC1, and LAMC2 were highly expressed (green) but not LAMB4 (red rectangle, Fig. S2C). Next, we assessed LAMB4 throughout mesoderm and cardiomyocyte differentiation (Fig. S2D). During the differentiation we measured high expression of classic mesoderm (TBXT, TBX6, and FOXF1, Fig. S2E) and cardiomyocyte markers (TNNT2 and NKX2–5, Fig. S2F), but not LAMB4. This was confirmed by analyzing published RNAseq data of human mesoderm and cardiomyocytes differentiated from hPSCs22. LAMA1, LAMB1, LAMB2, and LAMC1 were highly expressed (green), but not LAMB4 (red rectangle) during the assessed timepoints (Fig. S2G). Thus, we next investigated LAMB4 expression in ectoderm, specifically in neural crest, by following a protocol to differentiate hPSCs into SNs using chemically defined conditions23,24 (Fig. 1B). In this protocol, SNs are differentiated by going through all the developmental stages observed in vivo23. We found that LAMB4 mRNA is expressed in day 12 NCCs differentiated from hPSC-ctr-H9 cells, and it peaked in the early stages of SN specification, by around day 20 in our protocol (Fig. 1C). In contrast, laminin β4 isolated from cell lysates and the ECM increased over time and peaked at later stages of SN development (day 40–50, Fig. 1D and E). It is possible that this increase is caused by laminin β4 still being assembled and secreted to the ECM although not transcribed at high rates. To test this, we measured laminin β4 levels in the ECM alone. To do this, we lysed and removed the cells using ammonium hydroxide and resuspended the undisturbed ECM in Laemmli buffer followed by analysis by immunoblot25. Similar to our previous results, laminin β4 signal increased over time, which suggests that indeed laminin β4 was being continuously secreted (Fig. 1F and G). Finally, we confirmed our results by immunofluorescence, where we found that NCCs and SNs express laminin β4 (Fig. 1H and I). Together, our data shows that LAMB4 is expressed in NCC lineages which include SNs.
Figure 1. LAMB4/laminin β4 is expressed in neural crest cells (NCCs) and sensory neurons (SNs).
A) Comparison of laminin β chains expressed in humans. B) Schematics of the NC and SN differentiation protocol. C) LAMB4 expression during SN differentiation. hPSC-ctr-H9 SNs were harvested at the indicated times and mRNA expression of LAMB4 was measured by RT-qPCR (n=3 biological replicates). D) Laminin β4 expression during SN development. Total protein was isolated from SNs differentiated from hPSC-ctr-H9 cells at the indicated times and immunoblotted for laminin β4 and actin. E) Signal intensity of immunoblots from D) was measured, quantified, and normalized to day 16 (n=3 biological replicates). F) Levels of laminin β4 in the ECM of SNs. hPSC-ctr-H9 SNs were harvested on the indicated days and the ECM was collected and immunoblotted for laminin β4. Plates were coated with laminin β1 and was used as a loading control. G) Signal intensity of immunoblots from F) was measured, quantified, and normalized to day 20 (n=4 biological replicates). H) Laminin β4 expression in NCCs. Day 12 NCCs differentiated from hPSC-ctr-H9 cells were fixed and stained for laminin β4, SOX10, and DAPI. I) Expression of laminin β4 in SNs. hPSC-ctr-H9 SNs were fixed on day 20 and stained for the laminin β4, TUJ1, and DAPI. For C), E), and G), one-way ANOVA followed by Dunnett’s multiple comparisons test. ns, non-significant, *p<0.05, **p<0.005, ****p<0.0001. Graphs show mean ± SEM.
LAMB4 is required for neural crest cell migration
Next, we asked what is the role LAMB4 plays in SN development. To address this question, we knocked-out LAMB4 in healthy hPSC-ctr-H9 cells using CRISPR/Cas9 (Fig. S3A). We identified a homozygous (LAMB4−/−) and a heterozygous (LAMB4+/−) clone by Sanger sequencing (Fig. S3B). Although there were no phenotypical differences in the hPSC colonies compared to the parental hPSC-ctr-H9 cell line (LAMB4+/+, Fig. S3C), laminin β4 levels at the mRNA and protein levels were reduced in LAMB4−/− and LAMB4+/− SNs (Fig. S3D–F). We then used these cell lines to ask whether LAMB4 is required for the development of NCCs. We found that LAMB4−/− and LAMB4+/− can still differentiate into NCCs (Fig. 2A), however loss of LAMB4 made the characteristic “ridges”, formed by accumulation of NCCs22, smaller by inspection by brightfield microscopy (Fig. 2A, red arrows and B). We first hypothesized that the reduced area was due to a decrease in the number of NCCs. We tested this by assessing the number SOX10+ NCCs by immunofluorescence. We indeed found a reduced number of large clusters of SOX10+ cells in the LAMB4−/− and LAMB4+/− cells, although there was a large number of single SOX10+ cells (Fig. 2C). To confirm our results, we quantified the number of cells expressing the migratory NCC marker CD49d (which correlates well with SOX10 at this developmental stage23,26,27) by flow cytometry analysis. There was no change in the number of CD49d+ cells in any of the lines (Fig. 2D), suggesting that the number of NCCs was not affected by LAMB4. During development, NCCs migrate and accumulate forming ganglia28. Because the number of NCCs did not change upon loss of LAMB4, but we saw a high number of individual SOX10+ cells by immunofluorescence (Fig. 2C), we hypothesized that loss of LAMB4 impairs NCC migration. We first performed a scratch assay to test the migration of LAMB4+/+, LAMB4+/−, and LAMB4−/− NCCs. LAMB4+/− and LAMB4−/− NCCs failed to migrate after 48 hours compared to LAMB4+/+ (Fig. 2E and F). To confirm this, we performed live-cell imaging to map the migration of NCCs. Agreeing with our previous results, LAMB4+/−, and LAMB4−/− cells migrated at a lower accumulated distance compared to the LAMB4+/+ control (Fig. 2G and H). Finally, we characterized the expression of NCC genes in LAMB4+/− and LAMB4−/− NCCs. We found that SOX10 expression was similar in all the cell lines (Fig. 2I), agreeing with our flow cytometry results (Fig. 2D). In contrast, expression of genes involved in SN specification such as P75NTR, NGN1, and NGN2 was significantly downregulated in LAMB4+/− and LAMB4−/− NCCs (Fig. 2I). Together, our results show that LAMB4 is necessary for NCC migration and for SN differentiation.
Figure 2. LAMB4/laminin β4 is required for NCC migration.
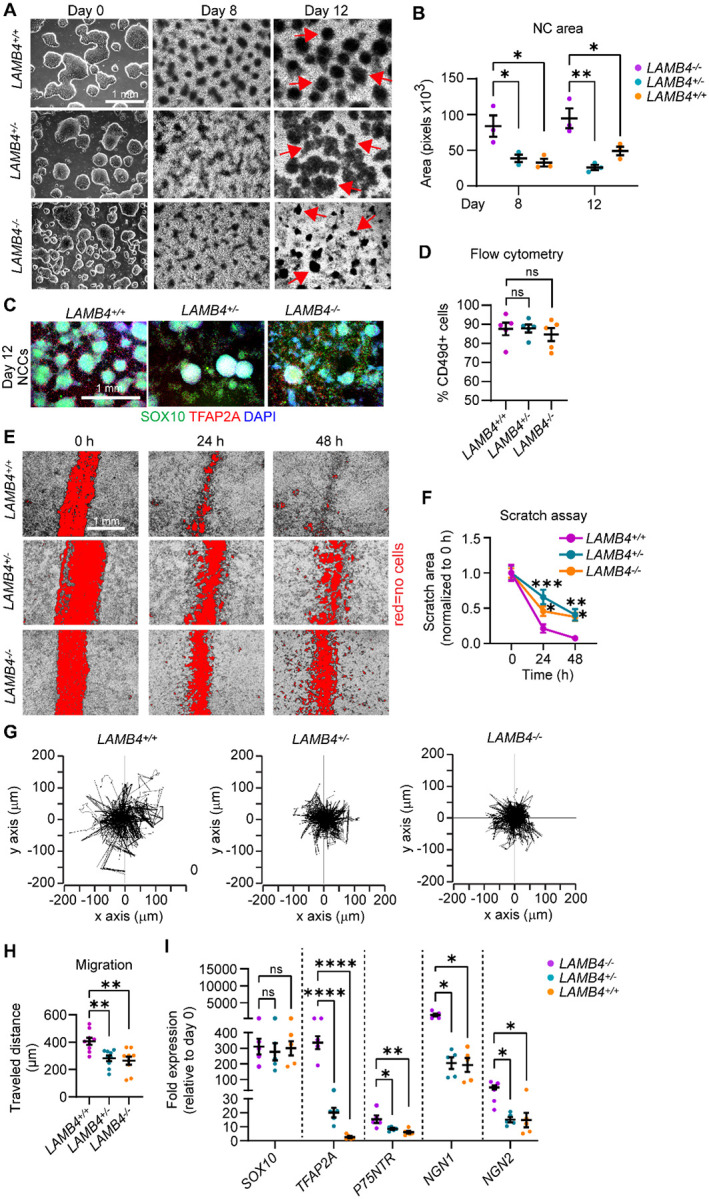
A) Effects of LAMB4 in NCCs. Brightfield images of colonies of LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs and NCCs at different stages. Red arrows indicate “ridges” of NCCs. B) Quantification of the area of NCC-ridges in A). Each dot indicates average of cell clusters that are part of the ridges (>70 clusters, n=3 biological replicates). C) Expression of NCC-markers upon loss of LAMB4. Representative immunofluorescence image of NCCs differentiated from LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs were fixed and stained for SOX10 (green), TFAP2A (red), and DAPI (blue). D) Number of NCCs differentiated from LAMB4 mutant hPSCs. NCCs differentiated from LAMB4+/+, LAMB4+/−, and LAMB4−/− cells were harvested, stained for the NCC surface marker CD49d, and analyzed using flow cytometry (n=5 biological replicates). E) Measurement of NCC migration by scratch assay. NCCs from LAMB4 mutant cells were replated on day 8 and scratched when they reached confluency. Brightfield images were taken at 0, 24, and 48 hours after the scratch to follow NC migration into the scratched surface (shown in red). F) Scratched areas in E) (red) were measured and normalized to 0 hours. Average of 5–10 wells per condition are plotted (n=3 biological replicates) G) Migration of NCCs by live-cell imaging. Day 8 NCCs from LAMB4+/+, LAMB4+/−, and LAMB4−/− cells were replated and imaged every 10 minutes for 18 hours. Individual cells were tracked, and their traveled distance and direction were measured and plotted. H) Accumulated distance traveled by NCCs measured in G). The average of 9 wells per condition are plotted (n=3 biological replicates). I) Expression of NCC-related genes upon loss of LAMB4. RNA from LAMB4+/+, LAMB4+/−, and LAMB4−/− NCCs (day 12) was isolated, and mRNA levels were measured using RT-qPCR (n=5 biological replicates). For B), D), H), one-way ANOVA followed by Tukey’s multiple comparisons test. For I), one-way ANOVA followed by Dunnett’s multiple comparisons test. For F), two-way ANOVA followed by Tukey’s multiple comparisons test. ns, non-significant, *p<0.05, **p<0.005, ****p<0.0001. Graphs show mean ± SEM.
LAMB4 is required for the development of sensory neurons
Our results suggest that LAMB4 plays an important role in directing NCCs into a SN fate. Thus, we asked whether loss of LAMB4 negatively affects the development of SNs in our human in vitro system. We found that on day 20 of our differentiation protocol, the number of neurons differentiated from LAMB4+/− and LAMB4−/− hPSCs was decreased (Fig. 3A). Moreover, by day 50, the size of the SN clusters, reminiscent to the ganglia observed in vivo29, were reduced in LAMB4+/− SNs and virtually inexistent in LAMB4−/− SNs (Fig. 3A). We confirmed these results by immunofluorescence, where the clusters of SNs, stained for the SN marker BRN3A and the pan-neuronal marker TUJ1, were reduced in LAMB4+/− and LAMB4−/− lines compared to the parental control (Fig. 3B and C). Additionally, the size of clusters and the number of SNs stained for ISL1+ (SN marker) and PRPH+ (peripheral neuron marker) were also reduced (Fig. 3B and D). When we quantified the number of SNs by flow cytometry we found that, when compared to wild type, both LAMB4+/− and LAMB4−/− were reduced, LAMB4 heterozygous hPSCs differentiate more efficiently than homozygous LAMB4 knockout cells (LAMB4−/−), suggesting that the LAMB4 expression level is important for SN development (Fig. 3E). These results showed that loss of LAMB4 impaired the development of SNs in vitro without changing the number of NCCs. We also found an increase in the number of non-neuronal cells expressing alpha-smooth muscle actin (αSMA) differentiated from LAMB4+/− and LAMB4−/− hPSCs compared to the parental control (Fig. S4A and B). Furthermore, ACTA2, the gene expressing αSMA, was upregulated in LAMB4+/− and LAMB4−/− cells (Fig. S4C). On the other hand, we didn’t see upregulation of genes expressed by sympathetic neurons (ASCL1), motor neurons (MNX1), enteric neurons (EDRNB), and other CNS cells (OLIG2, Fig. S4C). Together our results show that in the absence of LAMB4, NCCs do not differentiate into SNs efficiently and the number non-neuronal αSMA+ cells increases. These results strengthen our hypothesis that LAMB4 is necessary for NCC and SN-specification, and in its absence, NCCs take a non-neuronal cell fate.
Figure 3. LAMB4 is necessary for SN development and survival.
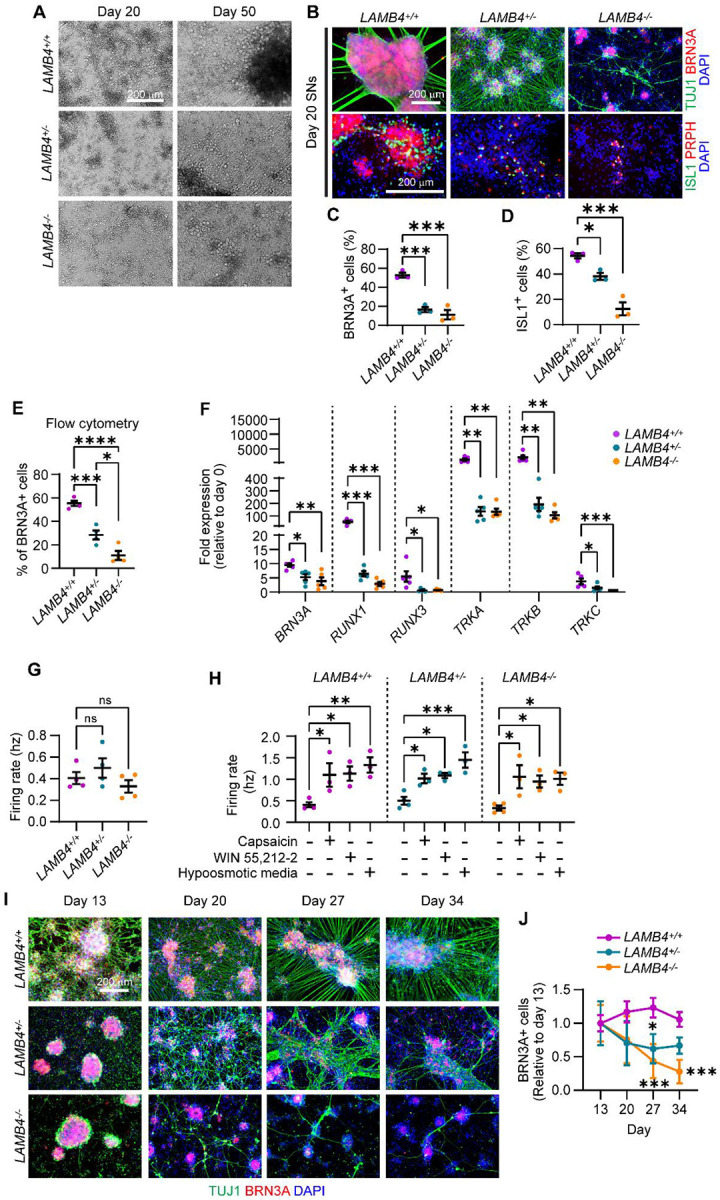
A) Effects of LAMB4 downregulation in SNs. Brightfield images of SNs differentiated from LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs at indicated days. B) Expression of SN markers upon loss of LAMB4. LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs were fixed on day 20 and stained for peripheral neuron markers (TUJ1 and PRPH) and SN markers (BRN3A, ISL1). Nuclei were stained with DAPI. C) Percentage of BRN3A+ cells in B). Normalized to DAPI. D) Percentage of ISL1+ cells in B). Normalized to DAPI. E) Quantification of the number of SNs differentiated from LAMB4 mutant hPSCs. SNs from LAMB4+/+, LAMB4+/−, and LAMB4−/− cells were harvested on day 20. SNs were then fixed, stained for BRN3A, and analyzed by flow cytometry (n=4 biological replicates). F) Expression of SN markers in LAMB4 mutant SNs. RNA from LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs was isolated on day 20 and gene expression was measured by RT-qPCR (n=5 biological replicates). G) Electrical activity of SNs upon loss of LAMB4. The firing rate of LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs was measured using multi-electrode array (MEA). Each dot represents the mean firing rate of 6 wells measured over 40 days (n=4 biological replicates). H) Electrophysiological changes of SNs to pharmacological nociceptor or mechanoreceptor activators. LAMB4-mutant SNs were incubated with nociceptor agonists (0.25 μM capsaicin and 1 μM WIN55,212–2) and a mechanoreceptor activator (hypoosmotic medium) and the electrical activity was measured using MEA. Each dot represents the mean firing rate of 6 wells measured over 40 days (n=4 biological replicates). I) Degeneration of LAMB4 mutant SNs. LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs were cultured in plates coated with fibronectin and poly-L-ornithine and reduced NGF concentration (1 ng/mL) and fixed on the indicated days. Cells were then stained for the neuronal marker TUJ1, the SN marker BRN3A, and DAPI. J) Quantification of BRN3A+ SNs from I) (n=3–4 biological replicates). For C), D), E), and G) one-way ANOVA followed by Tukey’s multiple comparisons test. For F) and H), one-way ANOVA followed by Dunnett’s multiple comparisons test. For J), two-way ANOVA followed by Šídák’s multiple comparisons test. ns, non-significant, *p<0.05, **p<0.005, ***p<0.001, ****p<0.0001. Graphs show mean ± SEM.
The three main subtypes of SNs found in the human dorsal root ganglia are nociceptors, mechanoreceptors, and proprioceptors, which detect pain, touch, and body position relative to space, respectively28. Since LAMB4 is necessary for SN development, we next asked whether its loss impacts a particular subtype or all of these SN types. During development, different genes are expressed to promote specification of SNs to a unique subtype. All SNs express BRN3A, whereas RUNX1 and TRKA are expressed by nociceptors during development. In contrast, mechanoreceptors express TRKB, and proprioceptors express TRKC. Additionally, progenitors of both mechanoreceptors and proprioceptors express RUNX328. We first analyzed the expression of these genes by RT-qPCR. BRN3A was downregulated in LAMB4+/− and LAMB4−/− SNs compared to LAMB4+/+ cells (Fig. 3F). Moreover, the nociceptor-related genes RUNX1 and TRKA were also downregulated, as well as RUNX3, TRKB, and TRKC, which are expressed by mechanoreceptors and proprioceptors (Fig. 3F). Additionally, the number of cells expressing TRKA, TRKB, and TRKC was reduced in LAMB4 mutant SNs measured by flow cytometry (Fig. S4D). Thus, LAMB4 is required for the development of all the three main SN subtypes. We next tested whether the neurons that developed were electrically active. We didn’t find any difference in the firing rate between LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs (Fig. 3G). Agreeing with this, the number, duration, frequency, and intervals of bursts were the same among the three lines (Fig. S4E–H). Furthermore, when we activated nociceptors with the agonists capsaicin and WIN55,212–223 the firing rate of LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs was similarly increased (Fig. 3F). This was also observed when LAMB4+/+, LAMB4+/−, and LAMB4−/− mechanoreceptors were activated using hypoosmotic medium23 (Fig. 3H), suggesting that LAMB4 does not play a role in SN function.
The ECM plays important roles in the homeostasis of different cell types, including neurons30. Thus, we asked whether LAMB4 is necessary for the survival of SNs. Our differentiation protocol had been optimized to assure the development and survival of wild type SNs31. To assess degeneration in non-wild type SNs, we first developed a modified protocol that accelerates degeneration in more vulnerable (for example diseased) lines, while still remaining robust cell survival in healthy SNs. This protocol consists of: (1) reducing the concentration of nerve growth factor (NGF) in the differentiation medium and (2) lack of coated laminin in the plates during the differentiation31. With this approach, we found that LAMB4+/− and LAMB4−/− SNs degenerate faster compared to LAMB4+/+ (Fig. 3I and J). However, LAMB4−/− SNs die at a faster rate compared to LAMB4+/−. Together, our results show that LAMB4 is required for both development and survival, but not function of SNs. Next, we sought to test whether altered LAMB4 expression has clinical implications.
A healthy extracellular matrix rescues the developmental phenotypes of the peripheral neuropathy Familial Dysautonomia
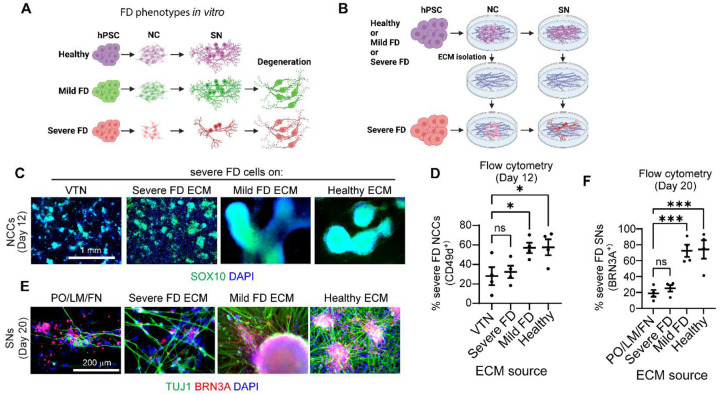
The peripheral neuropathy Familial Dysautonomia (FD) is a devastating genetic disease that specifically targets peripheral neurons32. It is caused by a mutation in the elongator complex scaffold protein ELP133,34. FD was one of the first diseases to be modeled using the iPSC technology35. While 99.5% of all FD patients harbor the precise ELP1 mutation, clinical symptoms vary widely among patients. The reasons for this discrepancy were unclear. To address this question, we reported that the severity of FD symptoms can be recapitulated in our in vitro system36. SNs differentiated from iPSCs of patients with mild symptoms showed only degenerative, but not developmental phenotypes. In contrast, iPSCs reprogrammed from patients with severe symptoms showed significant neurodevelopmental impairment, as well as neurodegeneration36 (Fig. 4A). We found that iPSCs from severe FD, but not mild FD patients harbored variants in LAMB4, which could account for the phenotypical differences36. Thus, we focused on addressing whether these variants in the FD patient iPSC lines affect LAMB4 expression. The ECM is important to maintain the availability of growth factors required for neuron development37. Therefore, loss of LAMB4 could affect the biophysical properties of the ECM and change the diffusion and availability of growth factors and signaling molecules. To confirm whether changes in the ECM affect the development of SNs in iPSCs derived from FD patients, we isolated the ECM of healthy cells as previously reported25. First, we differentiated healthy hPSC-ctr-H9 cells into NCCs and SNs, followed by lysis of the cells to maintain the healthy ECM. Severe FD iPSCs (iPSC-FD-S3) were then differentiated on top of the isolated healthy ECM (Fig. 4B). ECM from hPSC-ctr-H9 NCCs was sufficient to increase the area of the SOX10+ ridges and the number of NCCs (Fig. 4C and D). We observed the same results when iPSC-FD-S3 NCCs were replated on ECM deposited by hPSC-ctr-H9 SNs. The number of iPSC-FD-S3 SNs increased as measured by immunofluorescence and flow cytometry (Fig. 4E and F). As controls, we also isolated ECM deposited by NCCs and SNs differentiated from iPSCs of FD patients with mild (iPSC-FD-M2) and severe (iPSC-FD-S3) symptoms. We found that ECM from iPSC-FD-M2, but not iPSC-FD-S3, rescued the phenotypes similar to hPSC-ctr-H9 ECM (Fig. 4C–F). These results could be explained by the fact that both iPSC-FD-M2 and hPSC-ctr-H9 cells express WT LAMB4, whereas iPSC-FD-S3 cells have a variant in LAMB436. Therefore, we demonstrate that a healthy ECM is critical for NCC and SN development.
Figure 4. Healthy ECM rescues the developmental defects of SNs from severe FD iPSCs.
A) Phenotypes observed in SNs differentiated from iPSCs from FD patients with mild and severe FD symptoms36. B) Schematic of the experimental approach. C) Effects of the ECM in NCC differentiation. iPSC-FD-S3 iPSCs were differentiated on vitronectin (VTN) alone or on the isolated ECM deposited by healthy, mild FD, or severe FD NCCs. NCCs were fixed on day 12 and stained for SOX10 and DAPI. D) Quantification of NCCs from C). NCCs were harvested and stained for the NCC marker CD49d and analyzed using flow cytometry (n=4 biological replicates). E) Effects of the ECM on SN differentiation. iPSC-FD-S3 hPSCs were differentiated in dishes coated with Poly-L-Ornithine (PO), laminin-111 (LM), and fibronectin (FN) or on isolated ECM from healthy, mild FD, or severe FD SNs. SNs were fixed on day 20 and stained for the neuronal marker TUJ1 and SN marker BRN3A. F) Quantification of SNs from E). SNs were resuspended and fixed on day 20. Followed by staining for BRN3A and analysis using flow cytometry (n=4 biological replicates). For D) and F), one-way ANOVA followed by Dunnett’s multiple comparisons test. ns, non-significant, *p<0.05, ****p<0.0001. Graphs show mean ± SEM.
LAMB4 is downregulated in sensory neurons affected by severe Familial Dysautonomia symptoms
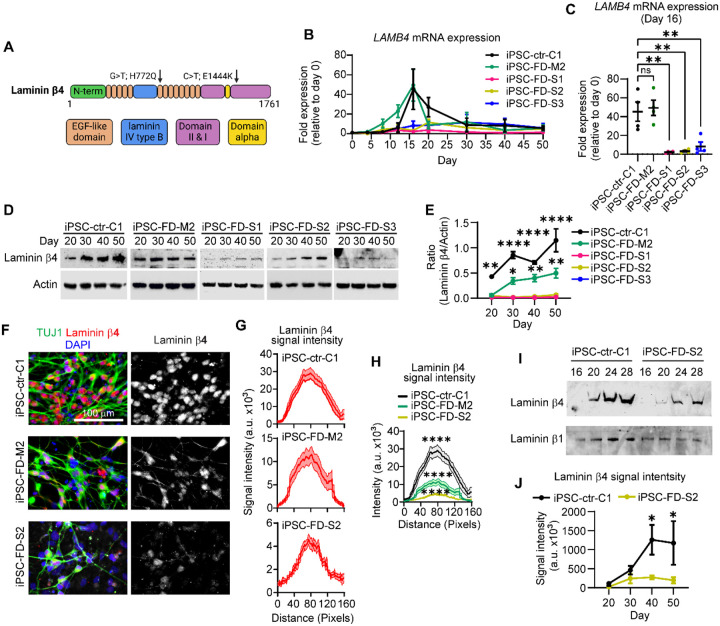
Our results suggest that the ECM, particularly LAMB4, plays a very important role in the development of SNs. Since two LAMB4 single nucleotide variants have been identified in patients with severe FD symptoms36 (Fig. 5A), we decided to investigate what are the consequences of these variants in LAMB4 expression. We used three iPSC lines from severe FD patients previously characterized36: iPSC-FD-S1, iPSC-FD-S2, and iPSC-FD-S3. As controls we used a healthy iPSC line (iPSC-ctr-C1) and an iPSC line derived from a patient with mild FD symptoms (iPSC-FD-M2), both of which do not harbor any variants in LAMB4. We first measured LAMB4 expression during development. Similarly to the control hPSC-ctr-H9 cells, iPSC-ctr-C1 and iPSC-FD-M2 cells expressed LAMB4 starting at the late stages of NCC differentiation and the early stages of SN specification (Fig. 5B). However, LAMB4 expression peaked on day 16, instead of day 20 in hPSC-ctr-H9 (Fig. 1D), possibly due to intrinsic differences between iPSCs and human embryonic stem cells (hPSC-ctr-H9). Interestingly, the three severe FD lines showed lower LAMB4 expression compared to the controls (Fig. 5B and C). We next tested whether transcriptional downregulation was also reflected at the protein level. We measured the expression of laminin β4 from cell lysates from SNs from day 20 to day 50 of the differentiation and found that it followed the same pattern as hPSC-ctr-H9 SNs. In the control line, low levels of laminin β4 were detected on day 20, which then increased until day 50, possibly due to the deposition of laminin β4 in the ECM (Fig. 5D and E). In contrast, laminin β4 levels did not increase in the severe FD SNs, suggesting that the variants observed in these lines affect LAMB4 transcription and subsequent translation (Fig. 5D and E). We confirmed these observations by immunofluorescence (Fig. 5F and G), where the signal intensity of laminin β4 in iPSC-ctr-C1 and iPSC-FD-M2 SNs was higher compared to iPSC-FD-S2 SNs (Fig. 5H). Finally, we measured the levels of laminin β4 in the ECM and confirmed that iPSC-FD-S2 SNs expressed lower laminin β4 levels compared to iPSC-ctr-C1 SNs (Fig. 5I and J).
Figure 5. LAMB4 expression is downregulated in patients with severe FD symptoms.
A) Schematic of the single nucleotide variants identified in LAMB4 in patients with severe FD36. B) LAMB4 expression in SNs differentiated from iPSCs of patients with severe FD. Severe FD iPSC lines S1, S2, and S3, one mild FD iPSC line (M2), and one healthy control iPSC line (C1) were differentiated into SNs. Total RNA was isolated in the indicated times and gene expression was measured by RT-qPCR (n=4 biological replicates). C) LAMB4 expression by SNs on day 16 shown in B) is shown (n=4 biological replicates). D) Laminin β4 expression during SN development. iPSC-FD-S1, iPSC-FD-S2, iPSC-FD-S3, iPSC-FD-M2, and iPSC-ctr-C1 cells were differentiated into SNs. Lysates were collected on the indicated days and immunoblotted for laminin β4 and actin. E) Quantification of signal intensity of immunoblots shown in D) (n=3 biological replicates). Difference between iPSC-FD-S1, iPSC-FD-S2, and iPSC-FD-S3 vs iPSC-ctr-C1 or iPSC-FD-M2 was analyzed. F) Laminin β4 in SNs. iPSC-FD-S2, iPSC-FD-M2, and iPSC-ctr-C1 cells were differentiated into SNs. Cells were fixed on day 20 and stained for laminin β4, TUJ1, and DAPI. G) Laminin β4 signal intensity measured from images in F). The average of 20 cells is plotted (n=3 biological replicates). H) Comparison of laminin β4 signal intensity from G). I) Laminin β4 levels in the ECM of severe FD SNs. ECM deposited by SNs from iPSC-FD-S2 and iPSC-ctr-C1 cells was isolated on the indicated days and immunoblotted for laminin β4. Plates were coated with laminin β1 which was used as a loading control. J) Signal intensity of blots from I) (n=4 biological replicates). For C), one-way ANOVA followed by Tukey’s multiple comparisons test. For E) and J), two-way ANOVA followed by Šídák’s multiple comparisons test. For H), two-way ANOVA followed by Tukey’s multiple comparisons tests. ns, non-significant, *p<0.05, **p<0.005, ****p<0.0001. Graphs show mean ± SEM.
We next asked whether restoring ELP1 expression rescues the expression of LAMB4 in FD. To answer this question we used two previously characterized FD iPSC lines (iPSC-rescued-T6.1 and iPSC-rescued-T6.5) where the ELP1 mutation was rescued36. However, since they were generated from iPSC-FD-S2 cells, they still harbor the LAMB4 variant identified in this cell line36. Similar to the parental line (iPSC-FD-S2), iPSC-rescued-T6.1 and iPSC-rescued-T6.5 cells expressed lower levels of LAMB4 mRNA compared to iPSC-ctr-C1 cells (Fig. S5A and B). This was confirmed by immunoblotting of the total levels of laminin β4 as well as immunofluorescence (Fig. S5C–E). Together, our results suggest that decreased LAMB4 expression in addition to the ELP1 mutation in FD may cause severe symptoms, and therefore has clinical implications. Moreover, LAMB4 could be used as a marker to detect early onset of severe symptomatology in FD and thus be used in personalized medicine.
Laminin β4 forms laminin-443 and controls actin filament accumulation in sensory neurons
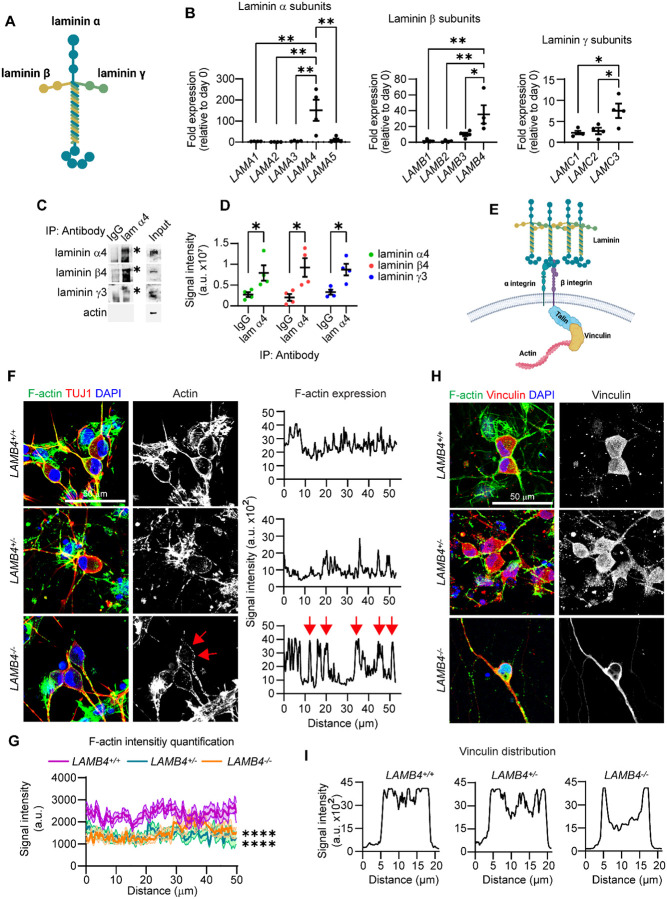
Since LAMB4/laminin β4 is necessary for SN development and has clinical implications, we decided to gain knowledge into how laminin β4 regulates SN development. Laminins are assembled in trimers consisting of chains α, β, and γ15 (Fig. 6A). There are five known α chains, four β chains, and three γ chains. To date, no laminin trimer containing laminin β4 has been described, thus, we first asked which chains interact with it. We measured the expression of every laminin chain in day 16 SNs and found that in addition to LAMB4, LAMA4 and LAMC3 were upregulated (Fig. 6B). LAMA4 encodes laminin α4 and LAMC3, laminin γ3, which suggests that laminin β4 is part of the laminin-443. To confirm this, we immunoprecipitated laminin α4 and found that laminin β4 and laminin γ3 came down as a complex (Fig. 6C and D). Next, we investigated the communication of the laminin β4-containing laminin trimer into the cell during SN development. Laminins bind to integrins at the plasma membrane, which through interactions with talin and vinculin, ultimately control the actin cytoskeleton (Fig. 6E). Through this mechanism, laminins ultimately regulate many cellular processes such as cell migration3. Based on the literature and our results showing that loss of LAMB4 affects NCC migration, we hypothesized that laminin β4 also controls actin in SNs. We looked at the expression of F-actin using confocal microscopy in SNs differentiated from LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs. F-actin signal was evenly distributed around the cell body of LAMB4+/+ SNs, however the expression changed into a slight punctuated pattern in LAMB4+/− SNs, and this pattern became more prominent in LAMB4−/− SNs (Fig. 6F). This change also correlated with a decrease in F-actin signal intensity (Fig. 6G), suggesting that laminin β4 regulates the formation of F-actin in SNs. Vinculin localization also changed upon loss of LAMB4 (Fig. 6H and I). Vinculin was detected throughout the cytoplasm of LAMB4+/+ SNs, possibly due to its localization at focal adhesions mediating the interaction between the cells and the ECM deposited on the surface of the well. In contrast, LAMB4+/− SNs showed vinculin accumulation at the plasma membrane in addition to the cytoplasm. Finally, cytoplasmic vinculin localization was lost in LAMB4−/− SNs and it was present mainly at the plasma membrane (Fig. 6H and I). These results show that the laminin β4, via laminin-443, maintains expression of F-actin at the cell body of SNs and transduces biophysical cues to the cell via vinculin. Moreover, in the absence of laminin β4, vinculin is no longer localized in the cytoplasm, potentially dissociated from focal adhesions and F-actin, thus affecting cell migration and differentiation. Our studies highlight a direct link between the cellular environment and intracellular components, and the critical role it plays during development which could potentially be targeted to treat peripheral neuropathies.
Figure 6. Laminin β4 interacts with laminin α4 and laminin γ3 and regulates actin filament (F-actin) expression.
A) Schematic of laminin trimer. B) Expression of laminin chains. RNA of day 16 hPSC-ctr-H9 SNs was isolated, and mRNA expression of all laminin chains was assessed by RT-qPCR (n=4 biological replicates). C) Immunoprecipitation of laminin α4. Lysates from hPSC-ctr-H9 SNs was collected on day 30, followed by immunoprecipitation of laminin α4 (lam α4) and immunoblotting for laminin α4, laminin β4, and laminin γ3. Asterisks mark the band corresponding for each protein. D) Quantification of signal intensity of immunoblots in C) (n=4 biological replicates). E) Schematic of regulation of intracellular pathways by laminins. F) Effects of LAMB4 downregulation on F-actin. LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs were fixed on day 20 and stained for F-actin (Phalloidin), TUJ1, and DAPI (left). Signal intensity of F-actin around the cell body from a representative experiment was measured and plotted (right). Red arrows indicate signal from actin puncta. G) F-Actin signal intensity of images from 20 cells in F) (n=3 biological replicates). H) Vinculin localization upon of loss LAMB4. LAMB4+/+, LAMB4+/−, and LAMB4−/− SNs were fixed on day 20 and stained for F-actin (Phalloidin), Vinculin, and DAPI. I) Signal intensity of vinculin from a representative experiment from H) was measured and plotted. For B), one-way ANOVA followed by Tukey’s multiple comparisons test. For D), two-tailed t-test. For G), two-way ANOVA followed by Tukey’s multiple comparisons test. *p<0.05, **p<0.005, ****p<0.0001. Graphs show mean ± SEM.
Discussion
LAMB4/Laminin β4 has been vastly understudied. We found LAMB4 orthologs in many species, including humans, chicken, zebrafish, and frogs, but not in rodents (Fig. S1A). In addition, we show that LAMB4 is expressed only in ectoderm lineages derived from NCCs and for a short period of time: 1) during the late stages of migratory NCC and 2) in early stages of SN specification (Fig. 1D). These factors make LAMB4 a difficult gene to study, as it cannot be studied in mouse models, and it is not widely expressed.
LAMB4 expression in late-stage NCCs and early-stage SNs suggests that it is important for the development of NCC-derived tissues including SNs. A report showing that laminin β4 is expressed in the cutaneous basement membrane38 and our results showing that LAMB4 downregulation reduces NCC migration (Fig. 2E–H) support this hypothesis. This timing also explains our gene expression results, where SOX10 expression in NCCs does not change in the absence of LAMB4. In contrast, genes expressed after SOX10 such as P75NTR, NGN1, and NGN2, which induce NCCs into SN lineages6,28, are downregulated. This is not unexpected, as other laminin chains also regulate NCC migration in vivo39. In SNs, accumulation of laminin β4 in the ECM is visible over 20 days after mRNA is downregulated. It is possible that the spike of LAMB4 transcription causes a burst of laminin β4 translation and secretion, which is necessary to further differentiate NCCs into SNs. The ECM is required for multiple aspects of development, maturation, and function of neurons30, including neurotransmission40, the activity of neuromodulators37, and promotes synaptogenesis41. Moreover, the ECM provides the necessary cues required for axon elongation and guidance during development and after injury30,42,43. We observed that axons of LAMB4−/− SNs show an irregular elongation pattern compared to control SNs (Fig. 3B), suggesting that laminin β4 is necessary for this process. These deficiencies in axon elongation could also affect SN homeostasis and explain why LAMB4−/− SNs degenerate faster than healthy SNs (Fig. 3I). This also suggests that laminin β4 secreted by differentiated SNs is required for their survival and agrees with the literature showing that neurons release laminins41,44–46.
LAMB4 downregulation is linked to diseases of the peripheral nervous system (peripheral neuropathies). Patients with sporadic cases of the enteric peripheral neuropathy diverticulitis have been shown to also harbor LAMB4 variants resulting in its downregulation16. Diverticulitis is caused by reduced neuronal density of NCC-derived enteric neurons47,48. LAMB4 has also been linked to the peripheral neuropathy FD. It was previously reported that patients with severe, but not mild FD symptoms harbor mutations in LAMB436. We show that SNs differentiated from iPSCs reprogramed from patients with severe FD had lower LAMB4 expression (mRNA and protein) compared to mild FD SNs (Fig. 5D–H). Approximately 99.5% of patients with FD have a mutation in ELP132, LAMB4 expression could explain the symptomatic differences between patients with mild and severe symptoms. Moreover, our results hint at the possibility that LAMB4 could be used as a diagnostic marker for severe FD onset early in life.
Restoring ELP1 expression in severe FD iPSCs did not impact LAMB4 expression, suggesting that LAMB4 expression is independent of ELP1. ELP1 is the scaffold protein of the elongator complex, and it is involved in transcription and tRNA modification during translation49. Our results showing that mild, but not severe FD SNs express high levels of LAMB4 further hints at the possibility that LAMB4 expression is not completely ELP1-dependent. Another possibility is that ELP1 primarily impacts laminin β4 translation due to defects in tRNA production. Thus, laminin β4 translation could be downregulated in mild FD SNs due to the ELP1 mutation, whereas in severe FD SNs, LAMB4 mRNA expression (due to the identified LAMB4 mutations) and translation (due to the ELP1 mutation) are both affected. Further studies will be necessary to dissect this mechanism.
Our studies show that laminin β4 is part of laminin-443. Laminin α4 has been shown to be expressed in the dorsal root ganglia (where NCCs further develop into SNs) during mouse development50 which strengthens the hypothesis that laminin β4 is involved in SN development. However, because LAMB4 is not expressed in rodents, it would be necessary to confirm this possibility in other in vivo models. Laminins bind to integrins located on the cell surface and connect to the actin cytoskeleton51. Laminin α4 has been shown to interact with integrin α3β1 and α6β152,53, however this interaction has not been explored in SNs. Our results suggest that laminin β4 activates integrins and cause the formation of F-actin around the cell body of SNs. Loss of LAMB4 results in reduced F-actin expression, possibly due to integrin inactivation. Interestingly, vinculin localization is also altered in LAMB4−/− SNs. Vinculin is a cytoplasmic protein that links integrins to actin and translates biophysical cues from the ECM into intracellular biochemical signals54. Upon integrin activation, vinculin is recruited to F-actin and focal adhesions51,54,55. Loss of LAMB4 resulted in changes in vinculin localization, possibly due to the inactivation of integrins. In this scenario, it is possible that vinculin associates with other known interactors present at the cell membrane, such as phosphatidylinositol 4,5-bisphosphate56. These are interesting observations, as most of the studies of actin in neurons focus on the growth cone57 and opens new avenues to study the cell biology of peripheral neurons. Furthermore, understanding ECM and actin regulation in the peripheral nervous system will uncover new mechanisms that could be used to promote neuronal regeneration and treat peripheral neuropathies.
Materials and methods:
hPSC maintenance
hPSC-ctr-H9 human embryonic stem cells (WA-09, WiCell) and all human induced pluripotent stem cells were grown at 37 °C with 5 % CO2 in vitronectin-coated dishes (ThermoFisher, cat# A31804, 5 μg/mL, 1 h at RT). Cells were fed daily with Essential 8 Medium + Supplement (Gibco, cat# A1517001). Cells were split at a 1:10 ratio using the following protocol: cells were washed with PBS, incubated with 0.5 mM EDTA, 3.08 M NaCl in PBS with for 2 minutes at 37 °C, and then resuspended in E8 + Supplement. iPSC-ctr-C1, iPSC-FD-M2, iPSC-FD-S1, iPSC-FD-S2, and iPSC-FD-S3 were previously characterized36.
Sensory neuron differentiation
Differentiation was done as previously described23,24. Prior to differentiation, plates were coated with vitronectin (5 μg/mL) and incubated for 1h at RT. On day of plating (day 0), hPSCs were washed with PBS, incubated with 0.5 mM EDTA, 3.08 M NaCl in PBS for 20 minutes, and plated at a density of 200,000 cells/cm2 in NC differentiation media (day 0–1) containing: Essential 6 Medium (Gibco, cat# A1516401), 10 μM SB431542 (R&D Systems, cat# 1614), 1 ng/mL BMP4 (R&D Systems, cat# 314-BP), 300 nM CHIR99021 (R&D Systems, cat# 4423), and 10 μM Y-27632 (Biogems, cat# 1293823). BMP4 concentration was titrated for each line. Accordingly, BMP4 was not used with iPSC-FD-S1 and iPSC-FD-S3 cells. The next day, the cells were fed with NC differentiation media (day 0–1). From day 2 to 12, cells were fed every two days with NC differentiation media (day 2–12) containing: Essential 6 Medium, 10 μM SB431542, 0.75 μM CHIR99021, 2.5 μM SU-5402 (Biogems, cat# 2159233), and 2.5 μM DAPT (R&D Systems, cat# 2634).
On day 10, plates were coated with 15 μg/ml poly-L-ornithine (PO, Sigma, cat# P3655) in PBS and incubated at 37 °C overnight. On day 11, the plates were washed 3X with PBS and coated with 2 μg/ml laminin-1 (LM, Cultrex, cat# 3401-010-02) and 2 μg/ml human fibronectin (FN, Corning, cat# 47743–654) in PBS and incubated overnight. On day 12, cells were resuspended using Accutase (Innovative Cell Technologies, cat# NC9464543) for 20 minutes, washed with PBS, and resuspended in SN Media containing Neurobasal media (Gibco, cat# 21103–049) containing 1X N2 (Gibco, cat# 17502–048), 1X B-27 (Gibco, cat# 12587–010), 2 mM L-glutamine (ThermoFisher, cat# 25030–081), 20 ng/ml GDNF (Peprotech, cat# 450–10), 20 ng/ml BDNF (R&D Systems, cat# 248-BD), 25 ng/ml NGF (Peprotech, cat# 450–01), 600 ng/ml of laminin-1, 600 ng/ml fibronectin, 1 μM DAPT and 0.125 μM retinoic acid (Sigma, cat# R2625). Cells were then replated at a density of 250,000 cells/cm2 onto PO/LM/FN coated plates. The media was replaced the following day. Cells were fed every 2–3 days. On day 20, DAPT was removed. Differentiation progress was followed using a brightfield microscope (Leica).
Endoderm differentiation
Endoderm differentiation was performed as described36,58,31,44. On day 0, hPSC-ctr-H9 cells were washed with PBS and incubated with Accutase for 20 min and seeded at a density of 100,000 cells/cm2 in RPMI medium (ThermoFisher, cat# 12633012) with Glutamax (ThermoFisher, cat# 35050061) and 100 ng/mL Activin A (R&D Systems, cat# 338-AC-010). Cells were fed daily for 3 days and FBS was added at increasingly concentrations: 0%, 0.2%, and 2%.
Mesoderm and cardiomyocyte differentiation
Cardiomyocyte differentiation was done as previously described59. hPSC-ctr-H9 colonies were washed with PBS followed by incubation with Accutase for 20 min. Cells were resuspended in E8 medium + supplement and seeded at a density of 100,000 cells/cm2. When the cells reached ~80% confluency, the cells were fed with RPMI medium supplemented with insulin-free B27 (ThermoFisher, cat# A1895601) and 6 μM CHIR99021 for 2 days. A day later, the media was replaced with RPMI + insulin-free B27. On day 4, cells were fed with RPMI + insulin-free B27 with 5 μM IWP2 (Cayman Chemical, cat# 13951). The following day, the media was replaced with RPMI + insulin-free B27. The cells were fed on day 7 with RPMI + insulin-free B27 and media was replaced every 2 days.
RNA isolation and RT-qPCR
RNA was isolated using Trizol (ThermoFisher, cat# 15596026) according to the manufacturer’s conditions and resuspended in 20 μL RNase-free water. RNA concentration and purity was measured using NanoDrop One (ThermoFisher). 1 μg of RNA was converted to cDNA using iScript cDNA Synthesis kit (BioRad, cat# 1708841) according to the manufacturer’s instructions and diluted 1:100 in RNase-free water. RT-qPCR reactions were run with 1 ul of cDNA and SYBR Green Supermix (BioRad, cat# 1725272) according to the manufacturer’s conditions in a C1000 Touch Thermal Cycler CFX96 (BioRad). The following cycling parameters were used: 95°C for 5 minutes, 40 cycles of 95°C for 5s and 60°C for 10 s. Results were analyzed using the comparative CT method. GAPDH was used as a housekeeping gene. The sequences of primers used in this study are available in Supplementary Table 1.
Antibodies
Laminin β4 (Abcam, cat# ab150819; Sigma, cat# HPA020242), laminin β1 (Abcam, cat# ab44941), laminin α4 (R&D Systems, cat# AF7340), laminin γ3 (Proteintech, cat# 67261–1-I), SOX10 (Santa Cruz, cat# sc-365692), TFAP2A (Abcam, cat# ab108311), BRN3A (Millipore, cat# MAB1585), TUJ1 (Biolegend, cat# 801201), ISL1 (DSHB, cat# 39.4D5-c), PRPH (Santa Cruz, cat# sc-377093), Actin (BD Biosciences, cat# 612656), Vinculin (Abclonal, cat# A14193), αSMA (Sigma, cat# A5228), Phalloidin-iFluor 488 (Abcam, cat# ab176753), CD49d-PE/Cy7 (Biolegend, cat# 304314), TRKA-PE (R&D Systems, cat# FAB1751P), TRKB-AF647 (R&D Systems, cat# FAB3971R), and TRKC-PE (R&D Systems, cat# FAB373P). The following secondary antibodies were used: From ThermoFisher: goat anti-mouse IgG1 AF488 (cat# A21121), goat anti-mouse IgG2a (cat# A-21131), goat anti-mouse IgG2b (cat# A21242), donkey anti-rabbit AF647 (cat# A31573), donkey anti-mouse AF488 (cat# A21202), goat anti-mouse HRP (cat# 62–6520), and goat anti-rabbit HRP (cat# 65–6120), Goat anti-rat HRP (cat# A18865). Donkey anti-sheep HRP antibody (Jackson Immunoresearch, cat# 713-035-003). The dilutions used are indicated in each section.
Immunoblotting
To collect cell lysates, cells differentiated in 6-well plates were washed with PBS and incubated with 120 μL of RIPA buffer (Sigma, cat# R0278) with 1 mM PMSF and 1X PhosSTOP (Roche, cat# 4906845001) for 15 minutes on ice. Cells were then scrapped and the lysate transferred to an Eppendorf tube, followed by mixing 10 s using a vortex and centrifuged at 12,000 RPM for 10 minutes at 4°C. Supernatants were transferred to a new Eppendorf tube and protein concentration was measured. Samples were mixed with 2X Laemmli buffer containing β-mercaptoethanol and ran in 7.5% polyacrylamide gels under denaturing conditions using MOPS buffer at 130 V. Proteins were transferred to a nitrocellulose membrane and blocked for 30 minutes in 5% non-fat dry milk in 0.1 % TBS-T (0.1% Tween-20, 50 mM Tris-HCl, 150 mM NaCl, pH7.6). Primary antibodies were added to the membranes in blocking buffer (laminin β4 – 1:1000, laminin α4 – 1:1000, laminin γ4 – 1:1000, Actin – 1:5000) and incubated overnight at 4 °C. Blots were then washed 3X with 0.1 % TBS-T and incubated with goat anti-mouse HRP, goat anti-rabbit HRP, goat anti-rat HRP, or donkey anti-sheep HRP antibody (1:5000) for 1 h at room temperature. Blots were washed 3X with 0.1% TBS-T and incubated with Clarity Western ECL Substrate (BioRad, cat# 1705061). Chemiluminescence signal was detected using UVP ChemStudio (Analytic Jena). Signal quantification was done using Image Studio Lite (LICOR).
Immunoprecipitation
Lysates were collected and concentration was measured as described above. Magnetic protein A/G beads (25 μL, ThermoFisher, cat# 88802) were pre-washed 3X with RIPA buffer with 1 mM PMSF and 1X PhosphoSTOP and incubated with 1 μg of laminin α4 antibody for 30 minutes at 4 °C in a rotator. Beads were then washed 3X with RIPA buffer with 1 mM PMSF and 1X PhosphoSTOP and incubated overnight with 1 mg of lysate. The following day, beads were washed 3X with RIPA buffer with 1 mM PMSF and 1X PhosphoSTOP, and resuspended in 2X Laemmli buffer.
Immunofluorescence
NCCs and SNs differentiated in 24- or 4-well plates were washed once with PBS and fixed with 4% paraformaldehyde (ThermoFisher, cat# AAJ19943K2) for 20 minutes at RT. Cells were then washed with PBS and incubated for 20 minutes with Permeabilization buffer containing 1% BSA, 0.3% Triton-X, 3% goat or donkey serum and 0.01% sodium azide in PBS. Cells were then incubated with the indicated primary antibodies (laminin β4 – 1:100, SOX10 – 1:100, TFAP2A – 1:500, BRN3A – 1:100, TUJ1 – 1:1500, ISL1 – 1:200, PRPH – 1:100, αSMA – 1:100) in Antibody buffer containing 1% BSA, 3% goat or donkey serum and 0.01% sodium azide overnight at 4°C. Cells were then washed 3X in PBS and incubated with secondary antibodies in Antibody buffer for 1 h. Cells were washed with PBS, incubated with DAPI (1:1,000) for 5 minutes, washed with PBS, and stored at 4°C. Imaging was done using a Lionheart FX fluorescence microscope (BioTek). Image analyses and quantifications were done in Fiji. For quantifications, 5 different fields were imaged and quantified. For confocal microscopy, 50,000 NCCs were seeded in PO/LM/FN-coated 4-well chamber slides (iBidi, cat# 80426) on day 12. On day 20, SNs were fixed and stained as described above. Primary antibodies used: TUJ1 – 1:1500, Vinculin – 1:100. Phalloidin-iFluor 488 (1:1000) was incubated with secondary antibodies for 1 h. Imaging was done in an Olympus FV1200 Confocal Laser Scanning Microscope using Argon and Helium-Neon lasers. Images were taken as Z-stacks of 3 μm of height. ImageJ was used to obtain maximum intensity projections and to measure the signal intensity profiles.
Flow cytometry
On the indicated days, cells were washed with PBS and incubated with Accutase for 30 minutes at 37 °C. Cells were then washed and resuspended in Flow buffer (DMEM, 2% FBS, and 1mM L-glutamine) followed by centrifugation at 200 g for 4 minutes. Cells were resuspended in cold PBS, counted, and diluted to a concentration of 1×106 cells/100μL. For NCCs, cells were centrifuged at 200 g for 4 minutes at 4 °C and resuspended in 100 μL of Flow buffer and incubated with CD49d-PE/Cy7 antibody (1:160) for 30 minutes, or with TRKA-PE (1:20), TRKB-AF647 (1:20), or TKC-PE (1:20) antibodies for 1 hour on ice. Samples were washed 2X with Flow buffer, resuspended in 300 μL of Flow buffer with DAPI (1:1000), filtered, and analyzed using a Cytoflex S (Beckman Coulter). For SNs, cells at a concentration of 1×106 cells/100μL were centrifuged, resuspended in 300 μL BD Cytofix buffer (BD Biosciences, cat# 554655), and incubated on ice for 30 minutes. Cells were centrifuged for 4 minutes at 2,000 RPM and resuspended in 600 μL of cold BD Perm/Wash buffer (BD Biosciences, cat# 554723). Goat serum (30 μL) was added to the cells and incubated on ice for 30 minutes. Cells were divided in 3 tubes (200 μL each): 1) unstained control, 2) secondary antibody control, and 3) sample. All tubes were centrifuged for 4 minutes at 2,000 RPM and the cells were resuspended in 200 μL of Antibody buffer (BD Perm/Wash buffer + 10 μL goat serum) with or without BRN3A antibody (1:100) and incubated overnight at 4°C. Cells were then washed twice with 300 μL BD Perm/Wash buffer, resuspended in Antibody buffer with or without AF488 goat-anti-mouse (1:500), and incubated on ice for 30 minutes. Cells were then washed 3X with BD perm/wash buffer, filtered, and analyzed using a Cytoflex S (Beckman Coulter). Analyses were done using FlowJo.
Scratch assay
On day 8, NCCs differentiated from LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs were washed with PBS and incubated with Accutase for 20 minutes at 37 °C. Cells were resuspended in NC differentiation media (day 2–12), counted, and replated at a density of 60,000 cells/cm2 in 4-well or 24-well plates. When the cells reached confluency, a scratch was performed in the center of the well using a 1 000 μL sterile tip. Brightfield images were immediately taken (0 h) was taken using a Lionheart FX (Bio-Tek) fluorescent microscope. Subsequent images were taken 24 and 48 h later at the same coordinates. Images were analyzed as previously described60.
Live-cell imaging
On day 8, NCCs from LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs were washed with PBS, incubated with Accutase for 20 minutes at 37 °C, and resuspended in NC differentiation media (day 2–12). Cells were then counted and replated at a density of 15,000 cells/cm2 in 4-well or 24-well plates. Medium was replaced the following day and brightfield images were taken every 10 minutes for 18 h using a Lionheart FX microscope (Bio-Tek) with climate control chamber. Cells were maintained at 37 °C with 5 % CO2 throughout the experiment. Each experiment was performed in triplicate (technical replicate) and approximately 60–80 cells were tracked per well. Individual images were compiled using Fiji and individual cells were tracked using TrackMate (v7.13.2)61,62. Tracks of individual cells were exported and analyzed using the Chemotaxis and Migration Tool software (Ibidi).
Generation of LAMB4 mutant hPSCs
Two gRNAs (GCTCAAGATGACTGCAACAG and CTGGTGATCTCCTGGTGGGC) targeting exon 3 of LAMB4 were selected using E-CRISP63 (available at www.E-CRISP.org). The oligos were annealed, phosphorylated, and ligated into PX458 using T4 DNA ligase. The resulting plasmid was transformed into DH5α bacteria and colonies were screened by sanger sequencing. The resulting plasmids (PX458-LAMB4gRNA1 and PX458-LAMB4gRNA2) were transfected into hPSC-ctr-H9 cells using Lipofectamine Stem Transfection Reagent (ThermoFisher, cat# STEM00001) following to the manufacturer’s protocol. After 48 h, cells were washed with PBS and incubated with Accutase for 20 minutes at 37 °C. The cells were transferred to a 15 mL conical tube, filled with PBS and centrifuged at 200 g for 5 minutes. The supernantant was aspirated and the pellet was resuspended in sorting medium containing Essential 8 Medium + Supplement, 1X
CloneR (Stemcell Technologies, cat# 05889), and 10 μM Y-27632. Cells were then counted and 2×106 cells were transferred to an Eppendorf tube and resuspended in 400 μL of sorting medium containing 0.4 μL of Propidium Iodide (ThermoFisher, Cat# P3566). The resuspended cells were filtered using a round-bottom FACS tube and GFP+ cells were sorted using a FACS Melody Cell Sorter System (BD Biosciences). Individual cells were sorted to VTN-coated 96-well plates with prewarmed 50 μL of sorting medium in each well. The cells were fed every 24 h for approximately 10 days. When colonies started to emerge, cells were transferred to 24-well plates using EDTA and the protocol previously described. Genomic DNA was isolated from each clone and screened. Positive clones were further expanded.
Electrophysiology experiments
Experiments were performed using a Maestro Pro (Axion Biosystems) multi-electrode array (MEA) system. On day 12, NCCs were seeded (250,000 cell/cm2) onto PO/LM/FN-coated BioCircuit MEA 96 plates (Axion Biosystems, cat# M768-BIO-96), containing 8 embedded electrodes/well, in SN Media as previously described, and allowed to continue differentiating. Recordings were made every 2–3 days at 37°C with a sampling frequency of 12.5 kHz for 5 minutes. Recordings from at least 6 wells per reading were averaged. Firing frequency was normalized to the number of active electrodes. Bursts were detected using Inter-Spike Interval. Capsaicin (Sigma, cat# M2028) and WIN 55,212–2 (R&D Systems, cat# 1038) were resuspended in DMSO and added to the cells 3 minutes prior to starting recordings. Hypoosmotic media was obtained by mixing SN Media with sterile water in a 45:55 ratio and it was added to the cells prior to recordings.
Degeneration assay
On day 12, NCCs from LAMB4+/+, LAMB4+/−, and LAMB4−/− hPSCs were replated on 4-well plates (ThermoScientific, cat# 12-565-72), at 250,000 cells/cm2, coated with PO/FN in SN media with 1 ng/ml NGF. Cells were fed every 2–3 days. DAPT was removed after day 20. Cells were fixed on day 13, 20, 27, and 34 and stained for BRN3A and TUJ1.
Extracellular matrix isolation and rescue experiments
NC- and SN-derived ECM was isolated as previously described25. To isolate ECM from SNs, day 12, hPSC-ctr-H9, iPSC-ctr-C1, and iPSC-FD-S2 NCCs were resuspended in Accutase as described above and seeded in 60 mm dishes. On day 30, cells were washed with 3 mL of PBS and incubated with 20 mM Ammonium Hydroxide (Sigma, cat# 221228–100ML-A). The dishes were constantly shaken for 5 minutes at RT, followed by 5 washes with 5 mL of de-ionized water. For immunoblotting, the ECM was scrapped and resuspended in Laemmli buffer containing β-mercaptoethanol and 100 mM dithiothreitol (DTT, RPI, cat# D11000) preheated heated at 95 °C for 2 min. For ECM rescue experiments, hPSC-ctr-H9, iPSC-FD-M2, and iPSC-FD-S3 were differentiated using the SN differentiation protocol described above. On day 12 (NCCs) and day 30 (SNs) the cells were treated following the ECM isolation protocol. The undisturbed ECM was kept in the plates in de-ionized water. To start the differentiation, water was aspirated and iPSC-FD-S3 cells were seeded following the SN differentiation protocol described above.
Bioinformatics
RNAseq data from endoderm21 (GSE52658) and mesoderm RNAseq22 (GSE85066) were analyzed. FPKM and TPM results were converted to log2 and graphed as heatmaps. For laminin chains analysis the following sequences from NCBI were used: 1) LAMB1: D. rerio (NP_775382), X. tropicalis (XP_002933140), M. musculus (XP_006515056), R. norvegicus (XP_003750185), C. lupus (XP_038279702), B. taurus (NP_001193448), M. mulatta (XP_014990159), H. sapiens (XP_047276315), P. troglodytes (XP_001165667), G. gallus (XP_046780211), A. carolinensis (XP_016849500), S. purpuratus (XP_030828530), D. melanogaster (NP_476618), A. mellifera (XP_006571829), C. elegans (NP_500734); 2) LAMB2: M. musculus (NP_001398157), R. norvegicus (XP_006243771), M. mulatta (XP_014986301), H. sapiens (XP_005265184), P. troglodytes (XP_016796574), C. lupus (XP_038283703), B. taurus (XP_010816035), G. gallus (NP_989497), A. carolinensis (XP_062829843), D. rerio (XP_005162102), X. tropicalis (XP_004914156), D. melanogaster (NP_524006); 3) LAMB3: D. rerio (XP_700808.6), G. gallus (XP_040547616), X. tropicalis (XP_012826649), A. carolinensis (XP_062834708), M. musculus (XP_006497296), R. norvegicus (XP_008768078), B. taurus (XP_005217424), C. lupus (XP_038526808), M. mulatta (XP_014973102), H. sapiens (XP_005273181), P. troglodytes (XP_054514183); 4) LAMB4: D. rerio (XP_068073408), X. tropicalis (XP_031754867), C. lupus (XP_038310194), M. mulatta (XP_028702003), H. sapiens (XP_011514277), P. troglodytes (XP_063672018), G. gallus (XP_040515061), A. carolinensis (XP_062837767). Alignments were done using Clustal Omega64 using default settings. The phylogenetic tree was visualized using Treeviewer.
Statistical analysis
All analyses and graphs were done using PRISM (GraphPad). Statistical analyses are indicated in each figure legends. Two-tailed Student’s t-test was used to compare two groups. One-way analysis of variance (ANOVA) followed by Dunnett’s or Tukey’s multiple comparisons test was used to compare three or more groups. Two-way ANOVA followed by Šídák’s multiple comparisons test was used to analyze data sets with two variables. Data presented are shown as mean ± SEM. In all experiments the differences were considered significant when p<0.05. The number of biological replicates (n) are defined as the number of independent differentiations started at least three days apart or from a different vial of cells. The number of biological replicates are indicated in the figure legends.
Supplementary Material
Acknowledgments:
We thank Dr. Yao Yao (University of South Florida), Dr. Michael Tiemeyer (University of Georgia), and Dr. Natalia Ivanova (University of Georgia) for their input in this project. We thank Dr. Abel Alcazar-Roman (Heinrich Heine University Düsseldorf) for critical reading of the manuscript. We also thank Julie Nelson from the CSRL Cytometry Shared Resource Laboratory (University of Georgia) for her help with flow cytometry experiments. Schematics were done using Biorender.com.
Funding:
This work was funded by the faculty start-up funds from the University of Georgia to N.Z. and NIH/NINDS 1R01NS114567-01A1 to N.Z.
Funding Statement
This work was funded by the faculty start-up funds from the University of Georgia to N.Z. and NIH/NINDS 1R01NS114567-01A1 to N.Z.
Footnotes
Declaration of interests: The methods to generate sensory neuron cultures are patented under PTC 17/555,581 (Zeltner and Saito-Diaz). All other authors declare no conflict of interest.
Data and material availability:
Requests for reagents should be directed to the corresponding author, Nadja Zeltner (nadja.zeltner@uga.edu)
References:
- 1.Frantz C., Stewart K. M. & Weaver V. M. The extracellular matrix at a glance. J Cell Sci 123, 4195–4200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphrey J. D., Dufresne E. R. & Schwartz M. A. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol 15, 802–812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnans C., Chou J. & Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15, 786–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozario T. & DeSimone D. W. The Extracellular Matrix In Development and Morphogenesis: A Dynamic View. Dev Biol 341, 126–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martik M. L. & Bronner M. E. Regulatory Logic Underlying Diversification of the Neural Crest. Trends Genet. 33, 715–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simões-Costa M. & Bronner M. E. Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen J. H., Coles E. G. & Wilkinson D. G. Molecular control of neural crest formation, migration and differentiation. Curr Opin Cell Biol 12, 719–724 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Mayor R. & Theveneau E. The neural crest. Development 140, 2247–2251 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y. et al. Matrix stiffness modulates the differentiation of neural crest stem cells in vivo. J Cell Physiol 234, 7569–7578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yap L., Tay H. G., Nguyen M. T. X., Tjin M. S. & Tryggvason K. Laminins in Cellular Differentiation. Trends Cell Biol. 29, 987–1000 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Klaffky E. et al. Trophoblast-specific expression and function of the integrin alpha 7 subunit in the peri-implantation mouse embryo. Dev Biol 239, 161–175 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Miner J. H., Li C., Mudd J. L., Go G. & Sutherland A. E. Compositional and structural requirements for laminin and basement membranes during mouse embryo implantation and gastrulation. Development 131, 2247–2256 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Pinzón-Duarte G., Daly G., Li Y. N., Koch M. & Brunken W. J. Defective formation of the inner limiting membrane in laminin beta2- and gamma3-null mice produces retinal dysplasia. Invest Ophthalmol Vis Sci 51, 1773–1782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner N. J. Integrins and the extracellular matrix: key mediators of development and regeneration of the sensory nervous system. Dev Neurobiol 71, 1054–1072 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Domogatskaya A., Rodin S. & Tryggvason K. Functional diversity of laminins. Annu. Rev. Cell Dev. Biol. 28, 523–553 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Coble J. L. et al. Identification of a rare LAMB4 variant associated with familial diverticulitis through exome sequencing. Hum. Mol. Genet. 26, 3212–3220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazin T. & Freed W. J. Human embryonic stem cells: derivation, culture, and differentiation: a review. Restor Neurol Neurosci 28, 589–603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joung J. et al. A transcription factor atlas of directed differentiation. Cell 186, 209–229.e26 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeltner N. & Studer L. Pluripotent stem cell-based disease modeling: current hurdles and future promise. Curr. Opin. Cell Biol. 37, 102–110 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Yao Y. Laminin: loss-of-function studies. Cellular and Molecular Life Sciences: CMLS 74, 1095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh K. M. et al. Efficient Endoderm Induction from Human Pluripotent Stem Cells by Logically Directing Signals Controlling Lineage Bifurcations. Cell Stem Cell 14, 237–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh K. M. et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166, 451–467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito-Diaz K., Street J. R., Ulrichs H. & Zeltner N. Derivation of Peripheral Nociceptive, Mechanoreceptive, and Proprioceptive Sensory Neurons from the same Culture of Human Pluripotent Stem Cells. Stem Cell Reports 16, 446–457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito-Diaz K. & Zeltner N. A protocol to differentiate nociceptors, mechanoreceptors, and proprioceptors from human pluripotent stem cells. STAR Protocols 3, 101187 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellewell A. L., Rosini S. & Adams J. C. A Rapid, Scalable Method for the Isolation, Functional Study, and Analysis of Cell-derived Extracellular Matrix. J Vis Exp (2017) doi: 10.3791/55051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai X. et al. SOX10 ablation severely impairs the generation of postmigratory neural crest from human pluripotent stem cells. Cell Death Dis 12, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fattahi F. et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 531, 105–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmigère F. & Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 8, 114–127 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Sleigh J. N., Weir G. A. & Schiavo G. A simple, step-by-step dissection protocol for the rapid isolation of mouse dorsal root ganglia. BMC Res Notes 9, 82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melrose J., Hayes A. J. & Bix G. The CNS/PNS Extracellular Matrix Provides Instructive Guidance Cues to Neural Cells and Neuroregulatory Proteins in Neural Development and Repair. Int J Mol Sci 22, 5583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito-Diaz K. et al. Genipin Crosslinks the Extracellular Matrix to Rescue Developmental and Degenerative Defects, and Accelerates Regeneration of Peripheral Neurons. bioRxiv 2023.03.22.533831 (2023) doi: 10.1101/2023.03.22.533831. [DOI] [Google Scholar]
- 32.González-Duarte A., Cotrina-Vidal M., Kaufmann H. & Norcliffe-Kaufmann L. Familial dysautonomia. Clin Auton Res 33, 269–280 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Slaugenhaupt S. A. et al. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum Mol Genet 13, 429–436 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Cuajungco M. P. et al. Tissue-Specific Reduction in Splicing Efficiency of IKBKAP Due to the Major Mutation Associated with Familial Dysautonomia. The American Journal of Human Genetics 72, 749–758 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee G. et al. Modeling Pathogenesis and Treatment of Familial Dysautonomia using Patient Specific iPSCs. Nature 461, 402–406 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeltner N. et al. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat. Med. 22, 1421–1427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelyshev Y. A., Kabdesh I. M. & Mukhamedshina Y. O. Extracellular Matrix in Neural Plasticity and Regeneration. Cell Mol Neurobiol 42, 647–664 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goletz S. et al. Laminin β4 is a constituent of the cutaneous basement membrane zone and additional autoantigen of anti-p200 pemphigoid. J Am Acad Dermatol 90, 790–797 (2024). [DOI] [PubMed] [Google Scholar]
- 39.Coles E. G., Gammill L. S., Miner J. H. & Bronner-Fraser M. Abnormalities in neural crest cell migration in laminin alpha5 mutant mice. Dev. Biol. 289, 218–228 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Nishimune H., Sanes J. R. & Carlson S. S. A synaptic laminin-calcium channel interaction organizes active zones in motor nerve terminals. Nature 432, 580–587 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Pyka M. et al. Chondroitin sulfate proteoglycans regulate astrocyte-dependent synaptogenesis and modulate synaptic activity in primary embryonic hippocampal neurons. European Journal of Neuroscience 33, 2187–2202 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Kubo T., Yamashita T., Yamaguchi A., Hosokawa K. & Tohyama M. Analysis of genes induced in peripheral nerve after axotomy using cDNA microarrays. J Neurochem 82, 1129–1136 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Roumazeilles L., Dokalis N., Kaulich E. & Lelievre V. It is all about the support — The role of the extracellular matrix in regenerating axon guidance. Cell Adhesion & Migration 12, 87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nirwane A. & Yao Y. Laminins and their receptors in the CNS. Biol Rev Camb Philos Soc 94, 283–306 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Hagg T., Portera-Cailliau C., Jucker M. & Engvall E. Laminins of the adult mammalian CNS; laminin-alpha2 (merosin M-) chain immunoreactivity is associated with neuronal processes. Brain Res 764, 17–27 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Omar M. H. et al. CNS Neurons Deposit Laminin α5 to Stabilize Synapses. Cell Rep 21, 1281–1292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wedel T. et al. Diverticular disease is associated with an enteric neuropathy as revealed by morphometric analysis. Neurogastroenterol Motil 22, 407–414, e93–94 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Lake J. I. & Heuckeroth R. O. Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G1–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen L., Humbert S., Saudou F. & Chariot A. Elongator – an emerging role in neurological disorders. Trends in Molecular Medicine 16, 1–6 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Miner J. H. et al. The laminin alpha chains: expression, developmental transitions, and chromosomal locations of alpha1-5, identification of heterotrimeric laminins 8–11, and cloning of a novel alpha3 isoform. J Cell Biol 137, 685–701 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouvard D., Pouwels J., De Franceschi N. & Ivaska J. Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat Rev Mol Cell Biol 14, 430–442 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Fujiwara H., Kikkawa Y., Sanzen N. & Sekiguchi K. Purification and characterization of human laminin-8. Laminin-8 stimulates cell adhesion and migration through alpha3beta1 and alpha6beta1 integrins. J Biol Chem 276, 17550–17558 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Pang X. et al. Targeting integrin pathways: mechanisms and advances in therapy. Sig Transduct Target Ther 8, 1–42 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atherton P., Stutchbury B., Jethwa D. & Ballestrem C. Mechanosensitive components of integrin adhesions: Role of vinculin. Experimental Cell Research 343, 21–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byron A. et al. A proteomic approach reveals integrin activation state-dependent control of microtubule cortical targeting. Nat Commun 6, 6135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakolitsa C., de Pereda J. M., Bagshaw C. R., Critchley D. R. & Liddington R. C. Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99, 603–613 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Omotade O. F., Pollitt S. L. & Zheng J. Q. Actin-Based Growth Cone Motility and Guidance. Mol Cell Neurosci 84, 4–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holloway E. M. et al. Differentiation of Human Intestinal Organoids with Endogenous Vascular Endothelial Cells. Dev Cell 54, 516–528.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang T. et al. 1-deoxysphingolipids bind to COUP-TF to modulate lymphatic and cardiac cell development. Dev Cell 56, 3128–3145.e15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pijuan J. et al. In vitro Cell Migration, Invasion, and Adhesion Assays: From Cell Imaging to Data Analysis. Frontiers in Cell and Developmental Biology 7, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tinevez J.-Y. et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Ershov D. et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat Methods 19, 829–832 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Heigwer F., Kerr G. & Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods 11, 122–123 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Madeira F. et al. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res 52, W521–W525 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for reagents should be directed to the corresponding author, Nadja Zeltner (nadja.zeltner@uga.edu)