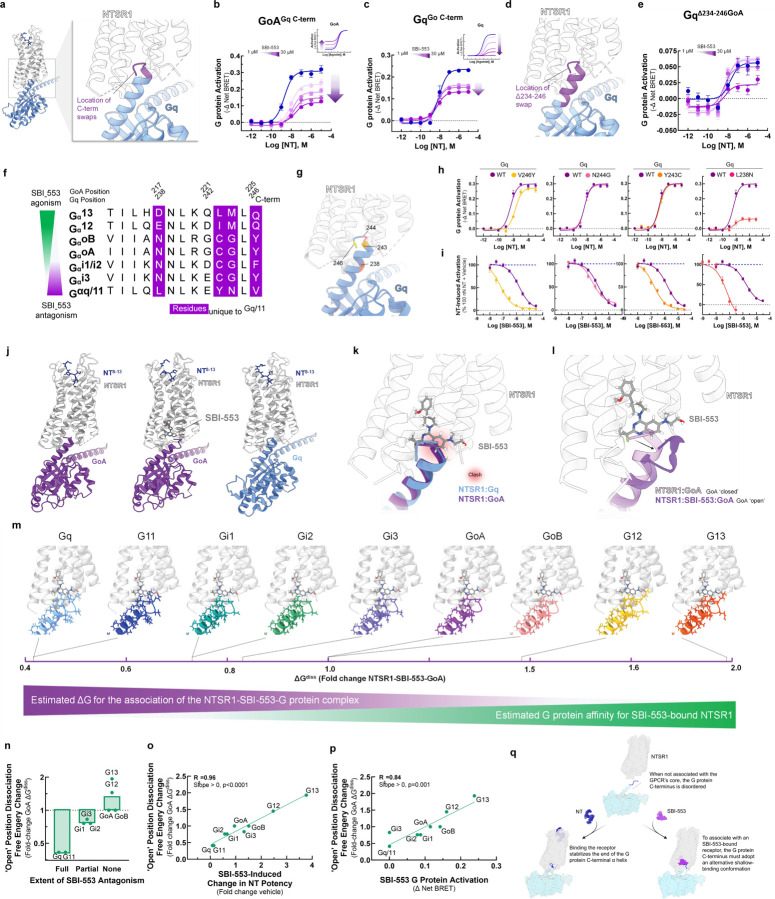
Figure 4. The sensitivity of a Gα protein to SBI-553 antagonism is determined by the ability of its C-terminus to adopt an alternative shallow-binding conformation.
(A-E) Sensitivity of G proteins to SBI-553 antagonism can be reversed by exchanging their C-termini. (A) Location of the GoA/Gq 5 C-terminal amino acid residue swap. (B) Swapping GoA’s 5 C-terminal amino acids for those of Gq confers sensitivity to SBI-553 antagonism. Inset, effect of SBI-553 on NT-induced activation of WT GoA for reference. (C) Swapping Gq’s 5 C-terminal amino acids for those of GoA reduces the antagonist efficacy of SBI-553. Inset, effect of SBI-553 on NT-induced activation of WT Gq for reference. (D) Location of Gq/GoA 13 C-terminal amino acid residue swap, GqΔ234-246. (E) Swapping Gq’s 13 C-terminal amino acids for those of GoA reduces the antagonist potency and efficacy of SBI-553. (F) Alignment of G protein C-termini. (G-I) Single amino acid substitutions on Gq’s C-terminus are insufficient to render Gq permissive of SBI-553. (G) Location of Gq point mutants on the NTSR1-Gq structure. (H) NT-induced activation of WT and mutant Gq constructs. (I) Effect of Gq mutagenesis on SBI-553’s ability to antagonize Gq activation by 100 nM NT. (J-L) Identification of an SBI-553-induced shallow-binding ‘open’ GoA conformation. (A) Cryo-EM structures showing NTSR1 bound by the NT active fragment and G protein in the absence and presence of SBI-553. (Left) NTSR1, NT, mini-GoA (PDB 8FN1). (Middle) NTSR1, NT, SBI-553, mini-GoA (PDB 8FN0). (Right) NTSR1, NT, mini-Gq (PDB 8FMZ). (K) Docking of SBI-553 in the 8FN1 and 8FMZ structures suggests that both GoA and Gq should clash with SBI-553. (L) Registration of the 8FN1 and 8FN0 structures illustrates that, in the presence of SBI-553, GoA adopts an alternative ‘open’ conformation to accommodate SBI-553 binding. (M-P) Homology modeling indicates that some NTSR1-SBI-553-’open’ position G protein complexes are more energetically favorable than others. In silico homology models were created based on the ‘open’ NTSR1-SBI-553-bound GoA conformation by changing the 13 C-terminal amino acids. Free energy of dissociation changes (ΔGdiss) were calculated, and models are positioned from most stable (right) to least stable (left). (N) NTSR1-SBI-553-G protein ‘open’ conformation ΔGdiss presented for individual G proteins, categorized by extent of sensitivity to SBI-553 antagonism. (O) Correlation of NTSR1-SBI-553-G protein ‘open’ conformation ΔGdiss with experimental SBI-553-induced fold change in NT G protein activation potency, as presented in Fig 2K. (P) Correlation of NTSR1-SBI-553-G protein ‘open’ conformation ΔGdiss with experimental max SBI-553-induced G protein activation, as presented in Fig 2D. (Q) Model of SBI-555 G protein subtype selectivity. For curve parameters, sample size, and statistical comparisons, see Table S4. For supporting data, see Figure S2, S3.