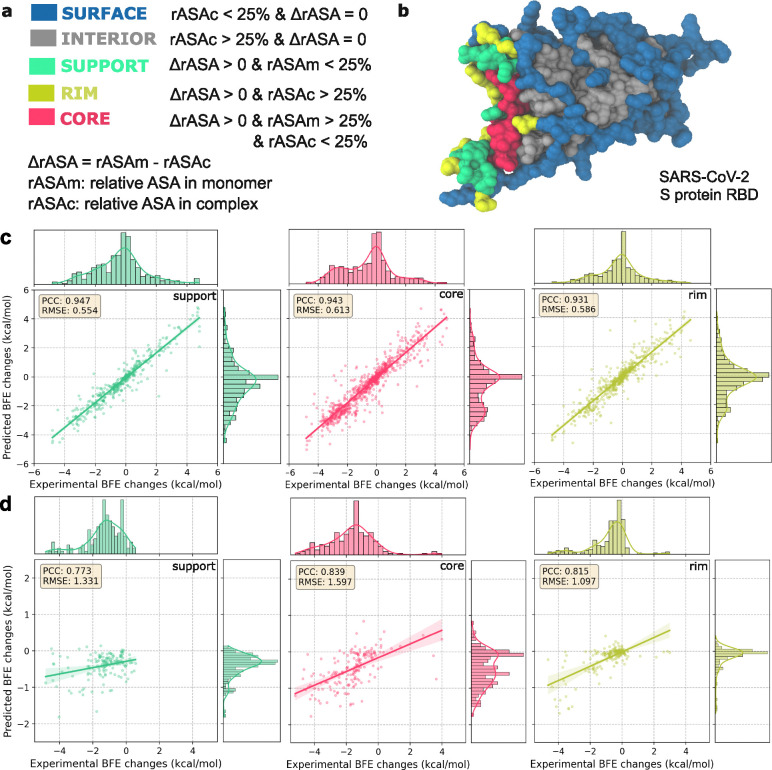
Figure 2:
(a): Definitions of structural regions based on relative ASA in monomer and complex [46]. (b): Structural regions on the Spike protein RBD (PDBID: 6M0J [47]). Amino acids are assigned to the surface, interior, support, rim, and core based on the rASA in monomer and complex. Structures are plotted by VMD [48]. (c): The 10-fold cross-validations for the four AF3-assisted SARS-CoV-2 RBD-ACE2 datasets displayed an average PCC of 0.933 and the average RMSE of 0.570 (see Figure 1(a)–(b)). The combined prediction results for different residue region types for four AF3-assisted SARS-CoV-2 RBD-ACE2 datasets according to Fig. 1(a) with PCCs of 0.947, 0.943, and 0.931 for the binding interfaces: support, core, and rim respectively. The average enrichment of the experimental DMS data is compared. (d): The 10-fold cross-validations of deep mutational scanning on the HK.3 RBD-ACE2[44] shows a PCC of 0.81 and a RMSE of 1.10. Prediction results for different residue region types according to Fig. 1(a) with PCCs of 0.773, 0.839, and 0.815 for the binding interfaces: support, core, and rim, respectively. The average enrichment of the experimental DMS data is compared.