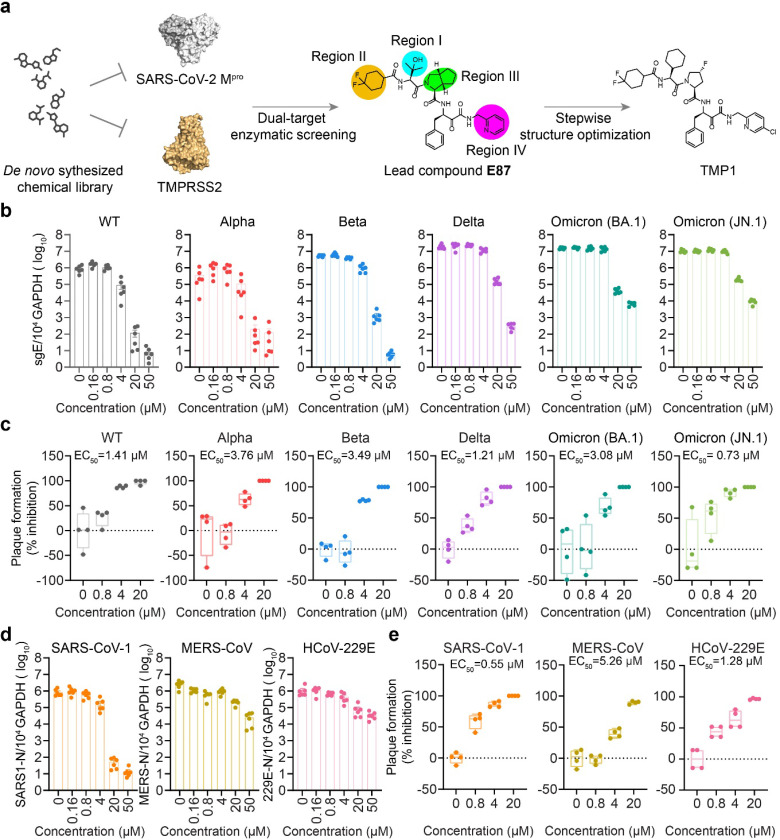
Figure 1. Discovery of the Mpro/TMPRSS2 bispecific inhibitor with highly potent anti-coronavirus efficacy.
(a) Schematic illustration of the screening workflow for the discovery of Mpro/TMPRSS2 bispecific inhibitor.
(b) Quantification of the subgenomic envelope (sgE) gene in VeroE6-TMPRSS2 cells (n=6) infected with wildtype SARS-CoV-2 and Alpha, Beta, Delta, Omicron (BA.1 and JN.1) variants in the presence or absence of TMP1. Lysates were harvested at 24 hpi. for one-step reverse transcription and quantitative polymerase chain reaction (RT-qPCR) analysis.
(c) Infectious viral titres in the supernatants harvested at 24 hpi. from VeroE6-TMPRSS2 cells (n=4) infected with wildtype SARS-CoV-2 and Alpha, Beta, Delta, Omicron (BA.1 and JN.1) variants were determined by plaque assays. Number of plaques were normalized to those recovered from supernatants with mock treatment only.
(d) Quantification of the nucleocapsid (N) gene in VeroE6-TMPRSS2 cells (n=6) infected with SARS-CoV-1 and MERS-CoV or in Huh7 cells infected with HCoV-229E at 24 hpi..
(e) Infectious viral titres in the supernatants harvested at 24 hpi. from VeroE6-TMPRSS2 cells (n=4) infected with SARS-CoV-1 and MERS-CoV or Huh7 infected with HCoV-229E were determined in VeroE6-TMPRRS2 (for SARS-CoV-1 and MERS-CoV) or Huh7 cells (for HCoV-229E) by plaque assays.
Each data point represents one biological repeat. Data represents mean ± SD from the indicated number of biological repeats. Data were obtained from two or three independent experiments. WT, wildtype SARS-CoV-2.