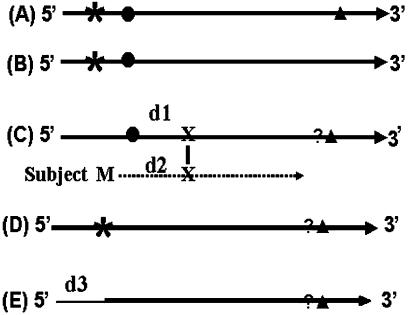
Figure 1.
Categories of algorithm-predicted cDNA clones. (A) A full-length sequence that includes one or more stop codons in the predicted 5′-UTR, a completely sequenced protein coding region and a 3′-UTR. (B) A sequence similar to those described in (A) except that the 3′ end of the ORF region is not sequenced. (C) A sequence having a start codon but lacking a stop codon in the 5′-UTR, whether it contains a potential translation start codon or not is determined by comparing the BLASTX alignment between its predicted protein and the subject. (D) A sequence having a stop codon in the 5′-UTR but lacking an in-frame start codon. This is an ambiguous sequence. (E) A sequence that includes a coding region but neither a stop codon nor a start codon in the sequenced portion. The length of the low quality sequence removed by Lucy (15) is taken into consideration when predicting whether or not it was a ‘possible full-length’ sequence. Asterisk: stop codon upstream of the start codon (5′ end stop codon); solid circle: predicted translation initiation codon; solid triangle: a stop codon downstream from the start codon (3′ end stop codon); question mark: indicates checking if a 3′ stop codon exists; (X): the first amino acid in the alignment of the HSP in BLASTX; (M): methionine; (d1) the length of predicted peptide from a predicted start codon to X; (d2) the length of M to X in the subject sequence of the HSP in BLASTX; (d3) length of EST sequence trimmed by Lucy, can include a portion of a vector, an adaptor and a low quality region of a cDNA sequence; thick solid line: sequences retained after processing by Lucy; thin solid line: the low quality sequence removed from the 5′ end by Lucy; dashed line: amino acid sequence of the subject in BLASTX.