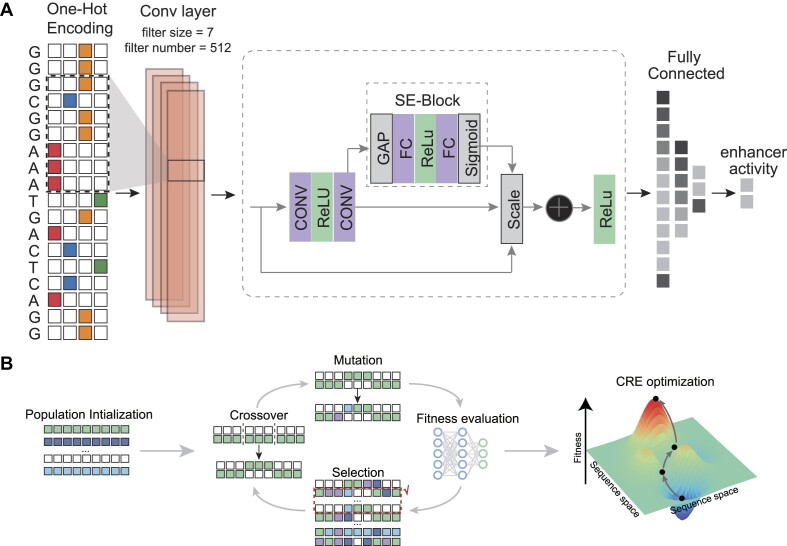
Figure 1.
Overview of the DREAM framework. DREAM comprises two integral modules: the state-of-the-art SENet that models enhancer activity using DNA sequences as the input (A) and the evolutionary optimization module of DNA sequences (B, ‘Genetic Algorithm’). The SENet was trained using UMI-STARR-seq data to learn the DNA regulatory lexicon underlying enhancer activity and was subsequently used to predict the regulatory activity of diverse DNA sequences. In tandem, the evolutionary optimization module employs a genetic algorithm, iteratively maximizing regulatory activity as predicted by the SENet-derived model. This iterative process ensures precise and targeted enhancement of enhancer functionality, thus facilitating the design of tailored sequences for specific regulatory tasks.