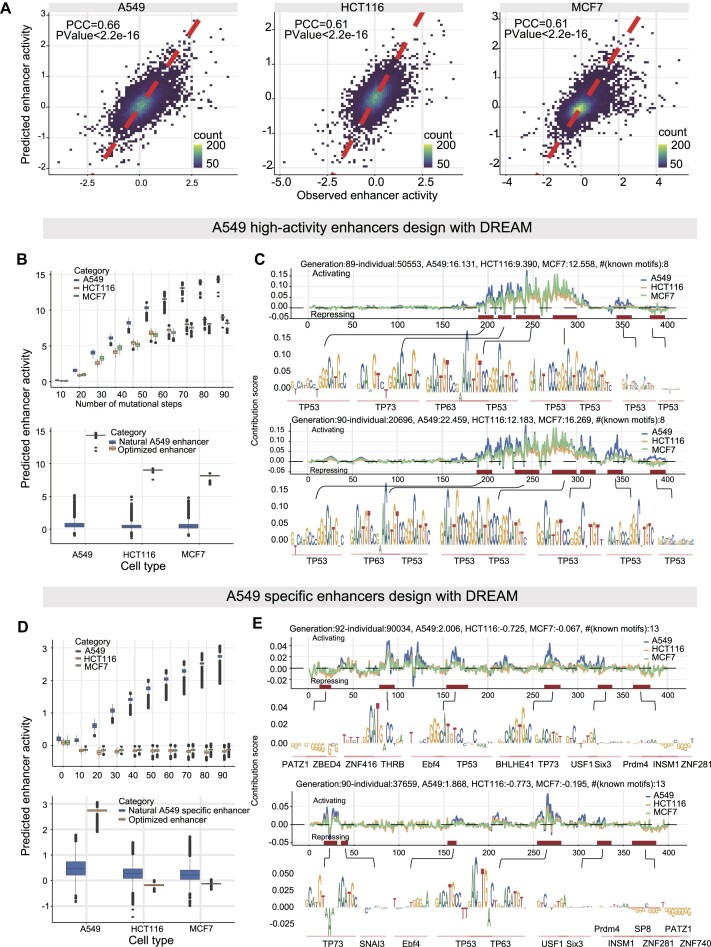
Figure 7.
A549 high-activity and A549 specific enhancers designed Using DREAM. (A) Predictive performance of the DREAM enhancer activity prediction module across A549, HCT116 and MCF7 cell lines, evaluated on the hold-out chromosome 22 test dataset. (B and D) Optimization processes for designing high-activity and cell-type specific enhancers in A549 cells, respectively. These figures also compare the enhancer activities of designed sequences to natural enhancers. (C and E) Nucleotide contribution scores of the optimized enhancer sequences, derived from the DREAM enhancer activity prediction module using DeepExplainer (62). Additionally, instances of motifs identified using Vertebrate PWMs from the JASPAR2024 database are shown. Boxes demarcate the 25th, 50th and 75th percentile values, while whiskers indicate the outermost point with 1.5 times the interquartile range from the edges of the boxes.