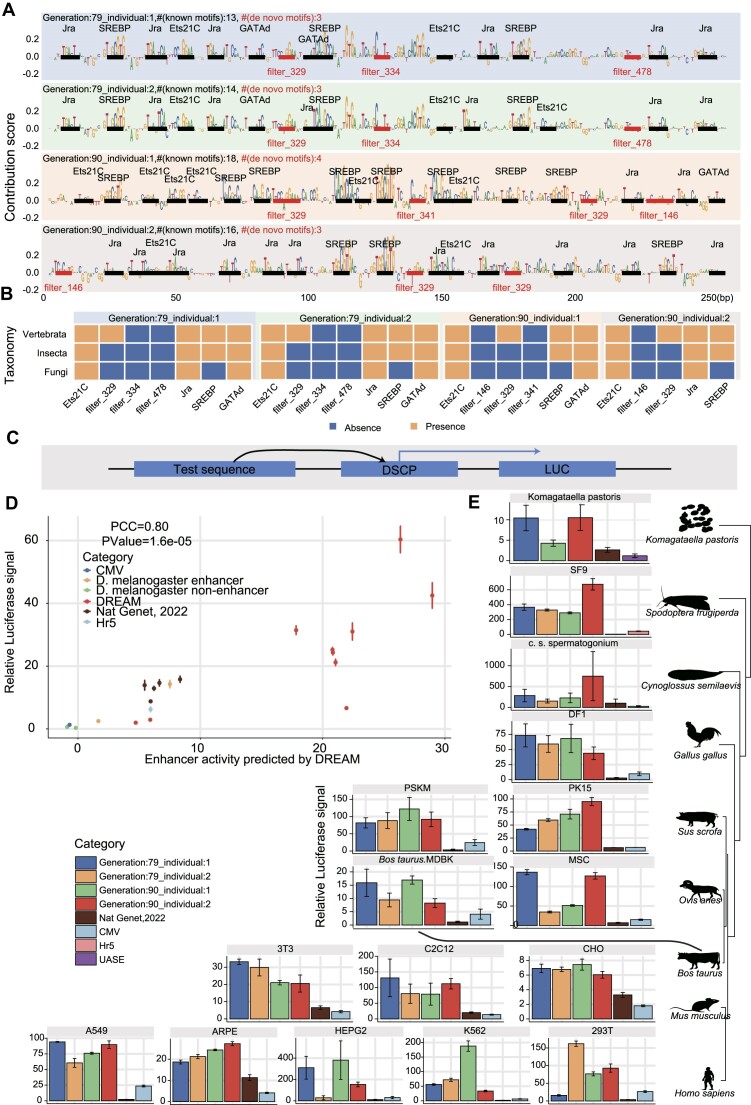
Figure 8.
The final optimized enhancers displayed extreme regulatory activity and are functionally conserved across diverse species. (A) Nucleotide contribution scores for the optimized enhancers derived from the enhancer activity models using DeepExplainer. Instances of motifs identified by DREAM are emphasized, with known motifs indicated in black and de novo motifs marked in red. The number of known motifs (# (known motifs)) and the number of de novo motifs (# (de novo motifs)) are also marked. (B) The colored matrices illustrate the presence or absence of TF motifs (x-axis) in the corresponding taxonomy (y-axis), with blue indicating absence and orange indicating presence of the TF in the respective taxonomy. (C) Schematic representation of the luciferase reporter assays system for the test sequences, with the designed sequences positioned at the 5′ end of the DSCP promoter. (D) Comparing enhancer activity, as measured by luciferase reporter assays, with predictions generated by the DREAM framework in S2 cells. Five classes of sequences are shown in the plot, including (i) non-enhancer sequences in the Drosophila genome (non-enhancer), (ii) the Drosophila developmental enhancers exhibiting the strongest and medium regulatory activity, (iii) the top five synthetic developmental enhancers with the highest regulatory activity, as designed by de Almeida et al. (35), (iv) ten enhancers designed by the DREAM design framework and (v) the CMV enhancer and Hr5 enhancer. The firefly luciferase values were normalized with the signal of Renilla luciferase. Error bars: Standard error of the mean (n = 3 biological replicates). (E) Comparative analysis using luciferase reporter assays to assess the activities of enhancers optimized by the DREAM framework, enhancers designed by de Almeida et al. (35), and the CMV enhancer across diverse cell lines spanning nine species, including chicken, fish (Cynoglossus semilaevis), human, mouse, pig, sheep, cattle, insect (Spodoptera frugiperda) and yeast (Komagataella phaffii). The Hr5 (81) and UASE (82) enhancers served as controls for insect (Spodoptera frugiperda) and yeast (Komagataella phaffii) cell lines, respectively, while the CMV enhancer was used as a control for the remaining cell lines. The luciferase values are normalized against the signal of Renilla luciferase. Error bars: Standard error of the mean (n = 3 biological replicates).