Figure 1.
Overview of gene expression in human milk
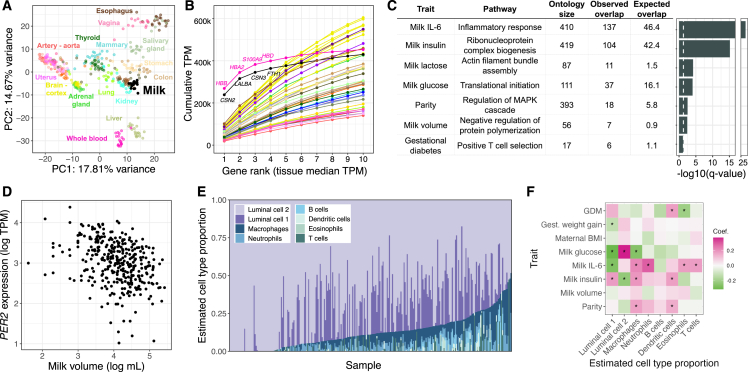
(A) Principal-component analysis of transcriptomes from a subset of GTEx tissues and milk. 19 random samples were chosen from each tissue. PCs were calculated using the 1,000 most variable genes within GTEx, and then milk samples were projected onto the GTEx samples. An equivalent plot including all GTEx tissues is shown in Figure S5.
(B) Cumulative TPM (transcripts per million) of the top 10 genes by median TPM for milk and GTEx tissues. The color scheme is the same as in (A).
(C) Gene Ontology enrichment of genes with expression correlated to maternal and milk traits. The most significant term for each trait is shown (STAR Methods). The dashed white vertical line denotes a q value of 0.05.
(D) Correlation between milk volume (from standardized electric breast pump expression during a study visit; STAR Methods) and PER2 gene expression in milk.
(E) Cell type proportion estimates generated using Bisque27 for transcriptomes from this study with reference milk single-cell RNA-seq from Nyquist et al.17
(F) Heatmap of regression coefficients between estimated cell type proportions (x axis) and maternal or milk traits (y axis) from a linear model including technical covariates (STAR Methods). ∗q < 10%.