Figure 2.
Genetic influences on gene expression in human milk
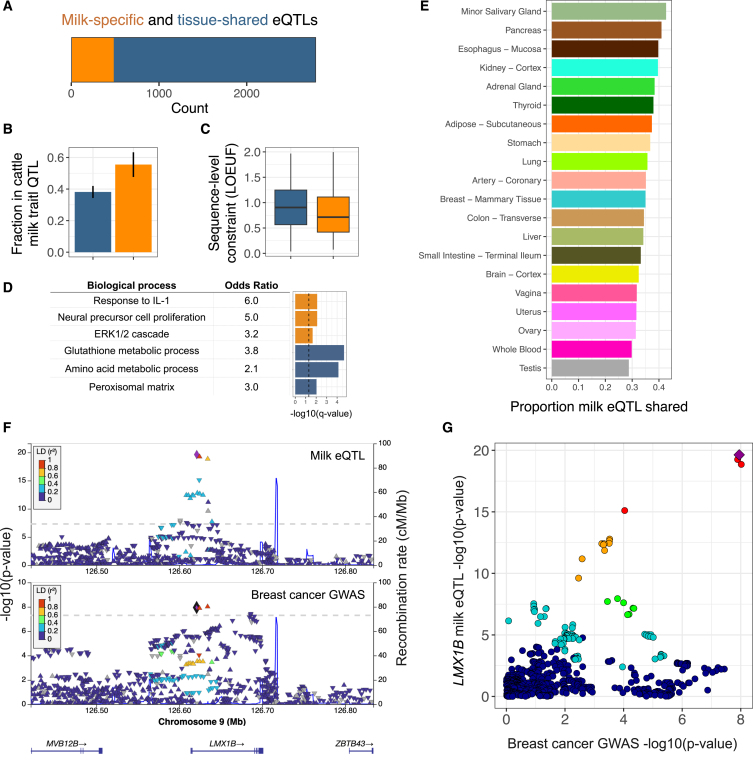
(A) Counts of genes with milk-specific eQTLs (orange, genes with an eQTL signal that did not colocalize with any GTEx tissue; STAR Methods) vs. tissue-shared eQTLs (blue, genes with all milk eQTL signals colocalized with at least one GTEx tissue).
(B) Fraction of genes in each category that overlapped with a milk trait QTL in the dairy cattle genome. Error bars represent a 95% confidence interval.
(C) Distributions of sequence-level constraint, measured by the loss-of-function observed/expected upper bound fraction statistic.38
(D) Enriched Gene Ontologies for genes with milk-specific (orange) or tissue-shared (blue) eQTLs. The dashed vertical line denotes a q value of 5%.
(E) Fraction of shared milk eQTLs with a subset of GTEx tissues, estimated with mash.40
(F) LocusZoom genetic associations in the LMX1B region with milk gene expression (top) and breast cancer risk (bottom). Each data point represents a SNP, plotted by its chromosomal location (x axis) and significance of association (y axis), with colors corresponding to linkage disequilibrium (r2) to the lead SNP for the milk eQTL, shown as a purple diamond.
(G) Each point is a variant, plotted by the strength of association with milk gene expression (y axis) and breast cancer risk (x axis). Colors are the same as in (F), top, with a purple diamond representing the lead milk eQTL SNP. The pattern of variants in the top right suggests a shared underlying causal variant.
See also Figures S13, S14, S15, S16, S17, S18, S19, and S20 and Tables S8, S9, S10, S11, S12, and S13.