Figure 3.
Effects of milk gene expression on HMO composition
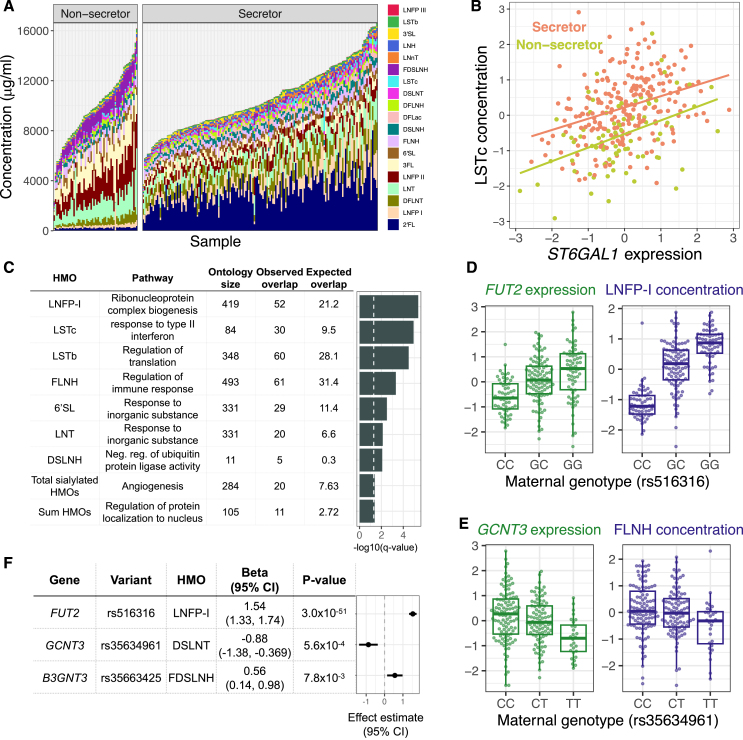
(A) HMO concentration (y axis) profiles for milk samples in our study (x axis), grouped by secretor status.
(B) Correlation between ST6GAL1 gene expression in milk and normalized LSTc concentration, colored by secretor status (log2 fold change = 0.32, p = 6.6 × 10−8, q = 1.5 × 10−4).
(C) Gene Ontology enrichment of genes with expression correlated to a single HMO or HMO category. The most significant term for each HMO is plotted. The dashed vertical line denotes a q value of 5%.
(D) Relationships between genotype at the lead SNP at the FUT2 eQTL and FUT2 expression in milk (green) or LNFP-I concentration (purple). LNFP-I concentrations are residuals after correcting for genetic PCs (STAR Methods).
(E) Relationships between genotype at the lead SNP at the GCNT3 eQTL and GCNT3 expression in milk (green) or FLNH concentration (purple). FLNH concentrations are residuals after correcting for secretor status and genetic PCs (STAR Methods).
(F) Estimates of the effect of milk gene expression of candidate HMO biosynthesis pathway genes on the abundance of HMOs from a Wald ratio test. Some genes had significant effects on more than one HMO (Table S18). The most significant HMO for each gene is plotted here.
See also Figures S21, S22, and S23 and Tables S14, S15, S16, S17, and S18.