Figure 4.
Interactions between milk gene expression and the infant fecal microbiome
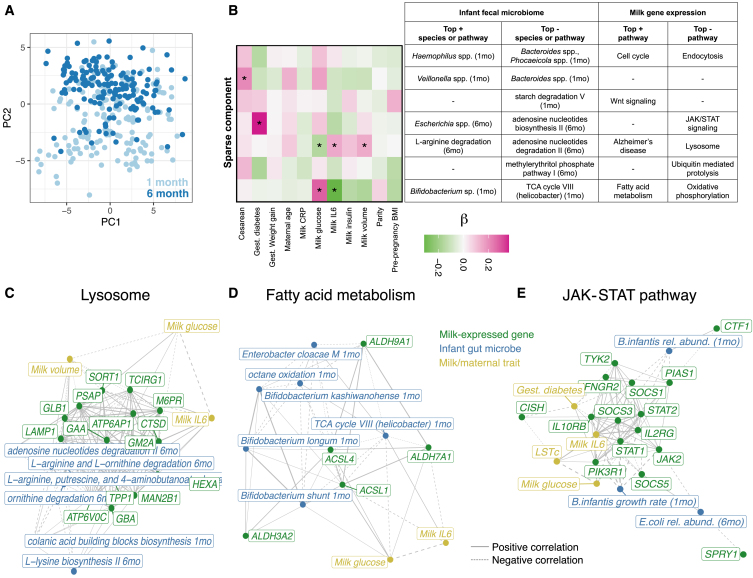
(A) Principal-component analysis of infant fecal microbiome metagenomic data, summarized at the taxonomic level, with each point representing a fecal sample and colors representing infant age (light blue, 1 month; dark blue, 6 months).
(B) Sparse CCA integrating milk host gene expression and infant fecal microbial species or microbial genetic pathway relative abundance (at 1 or 6 months of age) identified seven significant sparse components (in rows). The heatmap on the left shows Spearman correlation coefficients between each mother/infant pair score for a given sparse component (rows) and maternal or milk traits (columns). The table lists the most highly weighted microbial taxon or genetic pathway and the most significantly enriched host gene set in milk gene expression. (+) or (−) indicates whether these features were positively or negatively weighted in the sparse component.
(C and D) Network diagrams generated using the correlation matrix of infant fecal microbial species/pathways and milk-expressed host genes within an enriched pathway for two of the sparse components in (B). Line size corresponds to the absolute value of the correlation coefficient, and line type corresponds to negative (dashed) or positive (solid) correlations. Node color signifies milk-expressed host genes (green), infant fecal microbial pathways/taxa (green), or maternal/milk traits (yellow). Plotted edges had correlation p < 0.05.
(E) Network diagram displaying correlations between milk IL-6 concentration, LSTc (HMO) concentration, JAK-STAT pathway genes expressed in milk, and B. infantis relative abundance and estimated growth rate in the infant gut at 1 month and Escherichia coli relative abundance at 6 months. JAK-STAT pathway genes were selected that had a significant correlation with B. infantis or E. coli abundance after multiple test correction (q < 10%).
See also Figure S24 and Tables S19, S20, and S21.