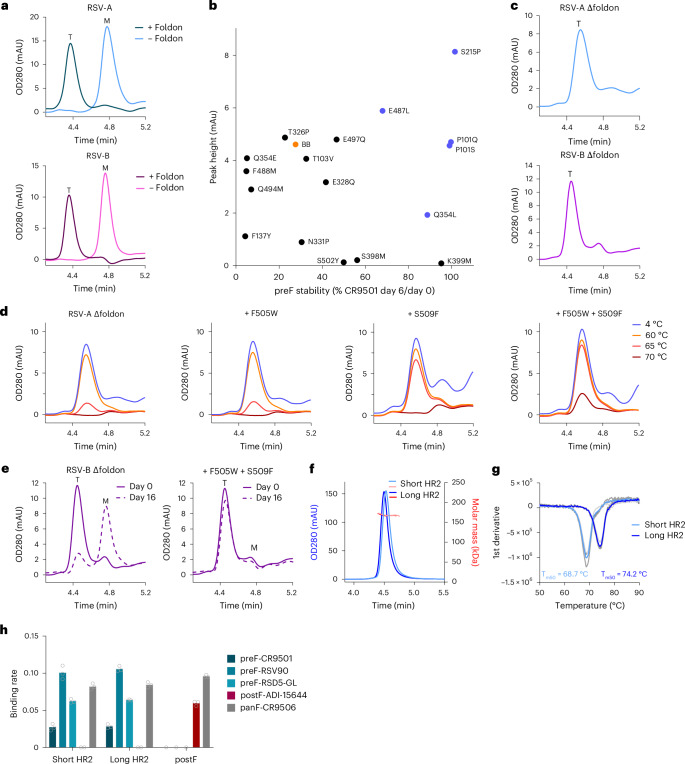
Fig. 4. Stable prefusion RSV F without a foldon domain through stabilization of the fusion peptide cavity and HR2 region.
a, Analytical SEC analysis of RSV-A and RSV-B F protein with or without foldon, as determined in cell supernatant. RSV-A F carries substitutions N67I, S215P and D486N. RSV-B F carries substitutions P101Q, I152M, L203I, S215P, D486N and D489Y. Trimeric (T) or monomeric (M) F is indicated. b, PreF expression and stability depicted as trimer peaks in analytical SEC at day 0 versus the CR9501 binding ratio at day 0 and 6. Stabilizing substitutions were screened in a semi-stable consensus backbone (‘BB’; RSV-A F with foldon and D486N). Substitutions in blue were selected for follow-up. c, Analytical SEC on RSV-A and RSV-B F without a foldon (Δfoldon) in cell supernatant. RSV-A Δfoldon contains P101Q, S215P, Q354L, D486N, E487L and D489Y. RSV-B Δfoldon contains P101Q, I152M, L203I, S215P, Q354L, D486N, E487L and D489Y. T is indicated. d, Stability of indicated variants as determined by analytical SEC on supernatants after 15 min of incubation at indicated temperatures. The backbone is RSV-A Δfoldon described in c. e, Stability of indicated variants as determined by analytical SEC on supernatants stored for 0 or 16 days at 4 °C. The backbone is RSV-B Δfoldon described in c. f, SEC-MALS (~162 kDa) of RSV-A F Δfoldon trimers with stabilizing substitutions P101Q, S215P, Q354L, D486N, E487L, D489Y, F505W and S509F. Variants contain either a short HR2 (residues 1–513) or a long HR2 (residues 1–524). g, Melting temperature (Tm50) of RSV-A F Δfoldon variants with a short or long HR2 sequence, as described in f, determined by differential scanning fluorimetry (DSF). Data reported as the average (blue line) of n = 3 technical replicates (grey lines). h, Antigenicity profile of RSV-A F Δfoldon variants with a short or long HR2, as described in f, measured by BLI. RSV postF protein is included as a control52. Antibody specificity is indicated. The binding rate is reported as the average of three technical replicates, and individual data points are represented by open circles.