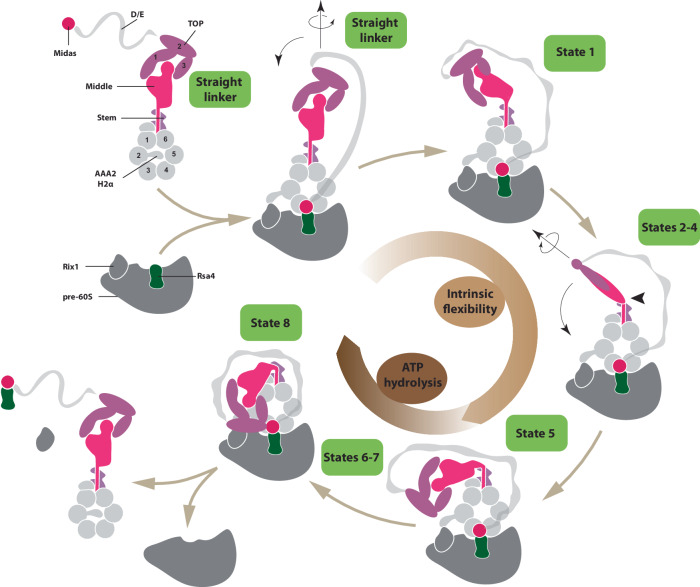
Fig. 8. Model for the Rea1 mechanism.
Rea1 in the absence of pre60S particles exists in an autoinhibited form with the AAA2H2α insert occupying the central pore of the AAA+ ring and the linker in the straight conformation. The binding of Rea1 to pre60S particles relocates AAA2H2α towards the pre60S particles, which allows the MIDAS domain to dock onto the AAA+ ring to engage with its assembly factor substrate (here Rsa4). The linker remains in the straight conformation. The linker subsequently rotates and pivots towards the plane of the AAA+ ring to reach state 1. From here the linker middle and top domains rotate around the long linker axis and swing towards the AAA+ ring. The region between the linker middle and stem domains acts as pivot point. In state 5, the linker middle and top domains are fully rotated and in close proximity to the AAA+ ring. In states 6-8, the linker top engages with the AAA+ ring and in the final remodelling step state 8 the linker top2/3 domains interact with the MIDAS domain to allow the transmission of force for assembly factor removal. Up to state 5, linker remodelling is nucleotide independent and driven by the intrinsic conformational flexibility of the linker. States 6–8 require ATP hydrolysis. Cartoon adapted from Sosnowski et al.17 Elife 7, 10.7554/eLife.39163 (2018) under a CC BY license: https://creativecommons.org/licenses/by/4.0/.