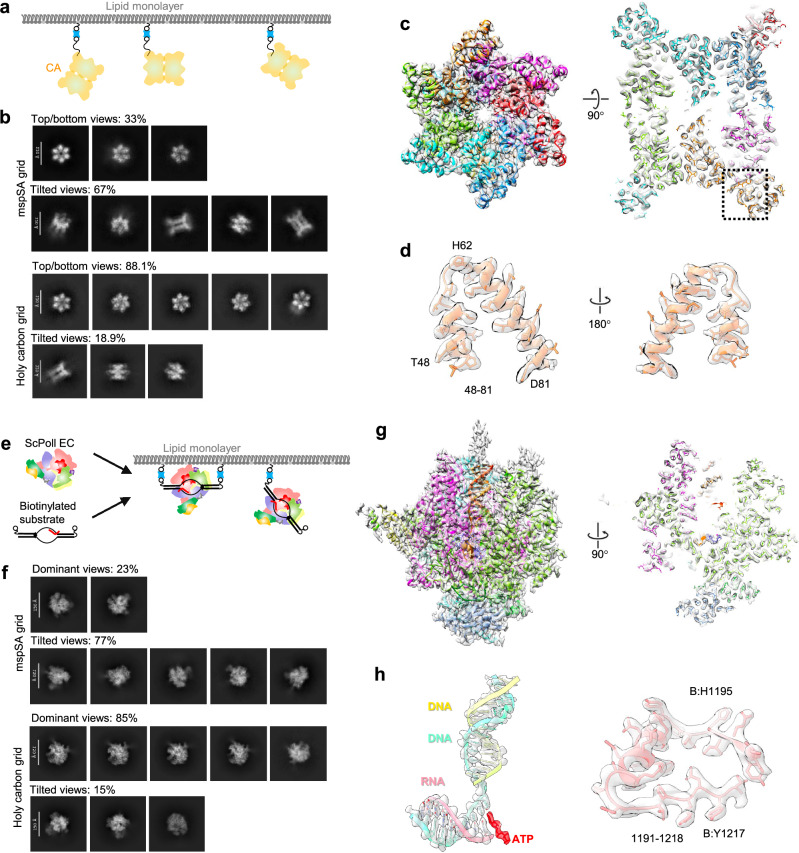
Fig. 5. CryoEM structures of HIV-1 CA Di-hexamer and scPol II EC complex.
a The design of mspSA affinity-capture of biotinylated HIV CA Di-hexamer. b Representative 2D classes of HIV-1 CA Di-hexamer generated from 1000 micrographs of mspSA affinity grid and holy carbon grid respectively applied same parameters for blob particle picking. 2D classes are separated into top/bottom- and tilted-views with indicated counts of each group. c Side views related to a 90° rotation of the cryoEM map of the HIV CA Di-hexamer at 3.14 Å resolution overlapped with the atomic models of subunits (colored). d Structural details of peripheral region of helix (aa: 48–81) in close-up views related to a 180° rotation. e The design of mspSA affinity-capture of biotin-tagged DNA/RNA in complex with scPol II. f Representative 2D classes of scPol II EC generated from 1500 micrographs of mspSA affinity grid and holy carbon grid respectively applied same parameters for blob particle picking. 2D classes are separated into dominant- and tilted-views with indicated counts of each group. g Side views related to a 90° rotation of the cryoEM map of the scPol II EC at 2.98 Å resolution overlapped with the atomic models of subunits (colored). h Structural details of peripheral regions of DNA/RNA and helix-loop conjunction aa: (1191–1218) in close-up views.