Abstract
Lumbar disc herniation (LDH) is a common degenerative disease of the lumbar spine, which is related to host genetic factors. Our study aimed to explore the association between MIR3142HG polymorphisms and LDH susceptibility. Six SNPs in MIR3142HG from 504 LDH patients and 500 healthy individuals were genotyped by the Agena MassARRAY platform. The relationship between SNPs and LDH susceptibility was evaluated with logistic regression analysis by calculating odds ratios (ORs) and 95% confidence intervals (CIs). The interactions between SNP and SNP were analyzed using the multifactor dimensionality reduction (MDR) method. Our study showed that rs7727115 was related to a decreased susceptibility to LDH. Rs2961920 and rs58747524 were significantly associated with an increased risk of LDH. Stratified analysis showed that rs7727115 reduced the risk of LDH in patients aged > 49 years. Rs17057846, rs2961920, and rs58747524 had a risk-increasing influence on patients aged > 49 years and women. Besides, rs7727115 decreased susceptibility in cases of disc prolapse, while rs2961920 and rs58747524 increased the risk. Rs2431689 increased susceptibility in patients with a single hernia, and rs58747524 correlated with an increased risk in cases of multiple hernias. Moreover, MDR analysis indicated that the combination of rs1582417, rs2431689, rs7727115, rs17057846, rs2961920, and rs58747524 was the best predictive model for LDH. Our study showed that MIR3142HG polymorphisms were significantly associated with LDH risk, which suggests that MIR3142HG polymorphisms play some potential roles in diagnosing LDH.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80758-8.
Keywords: Lumbar disc herniation, Genetic risk, MIR3142HG, Single nucleotide polymorphisms, Stratified analysis
Subject terms: Genetics, Molecular medicine, Risk factors
Introduction
Lumbar disc herniation (LDH) is a common lumbar degenerative disease caused by degeneration and displacement of the nucleus pulposus or the annulus fibrosus outside the intervertebral disc space 1. Pains in the leg or lower back are the main clinical features of LDH, which affect approximately 90% of the world’s population at some point during their life 2. At present, surgery is the most common treatment for LDH, but a large number of patients still suffer persistent low back and leg pain after surgical treatment 3,4. Thus, it is necessary to find new therapeutic targets for LDH. Several studies have shown that LDH is related to various inflammatory and immune responses. Among them, hematological indicators such as the neutrophil ratio (NEU), mean platelet volume (MPV), mononuclear cell ratio (MON), lymphocyte count (LYM), red blood cell distribution width (RDW), indirect bilirubin (IBIL) are closely related to the inflammation level and immune response of patients with LDH 5,6. As a common and multifactorial spinal disease, LDH is affected by multifactorial interactions, such as age, gender, height, smoking habits, trauma, and cold or humid environment as well as genetic factors. It has been reported that genetic factors such as gene polymorphisms play a more important role than environmental factors in the progression of LDH 7. To date, many susceptibility genes such as Storkhead Box1 (STOX1), CollagenType IX (COL9), Metallopeptidase Inhibitor 1 (TIMP1), Interleukin 4 (IL4), and Interleukin 6 (IL6), and MIR31HG polymorphisms were identified to be associated with LDH 8–10. However, the specific pathogenesis of LDH is still unclear.
Long non-coding RNAs (lncRNAs), non-coding transcripts of longer than 200 nucleotides, participate in the regulation of crucial biological processes. Growing evidence has indicated that lncRNAs are involved in numerous diseases, including intervertebral disc degeneration, and osteoarthritis 11–13. MIR3142HG, located at chromosome 5q33.3, can influence the transcription of MIR146A (the host gene of miR-146a), and then regulate the expression of miR-146a 14. MIR3142HG is involved in the occurrence of a variety of human diseases by affecting the inflammatory reaction, including lipopolysaccharide-induced acute lung injury and idiopathic pulmonary fibrosis 14,15. Previous studies have shown that inflammatory response plays an important role in the occurrence and development of intervertebral disc degeneration 16. There is evidence revealed that MIR3142HG can directly target miR-146a to regulate the inflammatory response in fibroblasts 14. Additionally, a recent study has shown that miR-146a can mediate the IL6/Signal Transducer And Activator Of Transcription 3 (STAT3) signaling pathway to regulate the occurrence of lumbar intervertebral disc degeneration 17. The above studies suggest that MIR3142HG has a crucial role in the occurrence of LDH. Single nucleotide polymorphisms (SNPs), the most common type of human heritable variants, are likely to be biomarkers for human disease 18. Several studies have indicated that MIR3142HG SNPs are significantly associated with human diseases. For instance, Cao et al. showed that rs17057846 and rs58747524 in MIR3142HG contributed to the elevated risk for IgA nephropathy 19. What’s more, a study has indicated that MIR3142HG rs1582417 and rs7727115 were significantly associated with osteonecrosis of the femoral head 20. However, the association between MIR3142HG genetic polymorphisms and LDH remains unclear.
Thus, we performed the case–control study (including 504 LDH patients and 500 healthy controls) to investigate the impact of MIR3142HG genetic variants on LDH susceptibility in the Chinese Han population. Our study will provide a new perspective on the molecular mechanism, prevention, and diagnosis of LDH.
Materials and methods
Study population
Before the study, G* Power (version 3.1.9.7) software was used to calculate the sample size. First, we selected the statistical method (t-test) and then chose classification (Difference between two independent means (two groups)). Then set the parameters: tail = 2, effective size = 0.2, α = 0.05, power = 0.884, and allocation ratio = 1. Finally, the calculated sample size of the case group and the control group was 499 cases respectively. A total of 1004 participants including 504 patients with LDH and 500 ethnicity, age, and gender-matched healthy controls were randomly recruited from the First Affiliated Hospital of Xi’an Jiaotong University. Patients were newly diagnosed as LDH by imaging examination and typical clinical symptoms and signs. Symptoms of LDH included: (1) partial lumbar pain and local typical sciatica; (2) low back pain; and (3) difficulty in straight leg elevation test and enhancement test. All controls were selected the healthy volunteers who were admitted to the hospital for physical examination at the same period as the cases and had no history of sciatica and low back pain. The controls showed to meet the following inclusion criteria: (1) subjects without any medical and family history of lumbosacral pain; (2) without any history of tumors; and (3) without spondylolisthesis, scoliosis, from trauma, osteoarthritis, rheumatism and rheumatoid arthritis. All participants with trauma, tumors, autoimmune diseases, and related lumbar diseases were excluded. We were informed about the purpose of the study and obtained an informed consent form from each subject before starting the study. The basic characteristics of each subject including age, gender, complication, blood indicators (NEU, (MPV), mononuclear MON, LYM, RDW, IBIL), degree, and level of herniation were obtained by medical records and standard questionnaires. This study was performed in accordance with the Helsinki Declaration and approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University.
SNP screening and DNA extracting
The SNPs in MIR3142HG were selected based on the following standards. First, we obtained the physical position of MIR3142HG on chromosome 5:160438594-160487426 by the human Ensembl database (release 110) 21. In the VCF to PED Converter window, we entered MIR3142HG's physical location, selected the Chinese Han population in Beijing (CHB) population, and downloaded the ped and info file for MIR3142HG SNPs. We obtained 103 SNPs within MIR3142HG from the database. Second, Haploview v4.2 software was performed for quality control (min genotype > 75%, Hardy–Weinberg equilibrium (HWE) > 0.05, minor allele frequency (MAF) > 5%, and r2 < 0.8). The linkage disequilibrium plot for the SNPs is shown in Figure S1. Finally, six candidate SNPs (rs1582417, rs2431689, rs7727115, rs17057846, rs2961920, and rs58747524) in the MIR3142HG gene were selected for investigation. The Gold Mag-Mini Purification Kit (Gold Mag Co. Ltd) was used to extract DNA from peripheral blood samples of study subjects. The extracted DNA was cryopreserved in ethylene diamine tetra acetic acid (EDTA) tubes. The DNA concentration was measured using NanoDrop2000, and the SNP was genotyped using Agena MassARRAY RS1000. The genotyping data was analyzed by Agena Typer Software (version 4.0). The primer sequences for PCR amplification are shown in Table S1.
Bioinformatic analysis
The possible functions of the six selected SNPs were predicted using HaploReg v4.2 online software (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). Additionally, we explored the effects of SNPs on the expression of MIR3142HG through the expression quantitative trait loci (eQTL) analysis of the Genotype-Tissue Expression (GTEx) database (http://www.gtexportal.org/).
Statistical analysis
SPSS 20.0 software was used to perform all statistical analyses. The t-test and chi-square test were used to assess the continuous variables (age, NEU, MPV, MON, LYM, RDW, and IBIL) and non-continuous variables (gender) between the case and control groups, respectively. The association between MIR3142HG polymorphisms and LDH susceptibility was determined by calculating 95% confidence interval (CI) and relative odds ratio (OR) with logistic regression analysis under multiple genetic models. The Benjamini and Hochberg’s false discovery rate (FDR) method was used to correct for multiple testing. The SNP-SNP interactions were analyzed by open-source Java software multifactor dimensionality reduction (MDR), version 3.0.2 22, and generalized multifactor dimensionality reduction (GMDR) (version 0.7). The one-way ANOVA method was used for index analysis.
Results
Characteristics of the study population
A total of 504 LDH patients (294 males and 210 females) and 500 healthy controls (293 males and 207 females) are included in this study (Table 1). The mean ages of cases and controls were 49.24 ± 14.88 years and 49.06 ± 14.47 years, respectively. There was no significant difference between the two groups in terms of gender (p = 0.642), age (p = 0.644), and RDW (p = 0.165). Statistically significant differences in NEU (p < 0.001), MPV (p < 0.001), MON (p = 0.042), LYM (p = 0.001), and IBIL (p < 0.001) were observed between the case and control groups. Additionally, the degree of herniation included disc herniation (110 cases), disc prolapse (388 cases), disc displacement (1 case), and others.
Table 1.
Basic characteristics of the LDH patients and the controls.
| Characteristics | Cases (n = 504) | Control (n = 500) | p-value |
|---|---|---|---|
| Age | 49.24 ± 14.88 | 49.06 ± 14.47 | 0.849a |
| ≤ 49 | 254(50%) | 248(50%) | |
| > 49 | 250(50%) | 252(50%) | |
| Gender | 0.949b | ||
| Male | 294(58%) | 293(59%) | |
| Female | 210(42%) | 207(41%) | |
| Complication | |||
| yes | 221(44%) | ||
| no | 283(56%) | ||
| NEU (%) | 68.93 ± 14.75 | 57.64 ± 8.44 | < 0.001a |
| MPV (fL) | 11.46 ± 1.36 | 10.85 ± 1.28 | < 0.001a |
| MON (%) | 6.69 ± 2.33 | 6.98 ± 1.95 | 0.042a |
| LYM (109/L) | 1.66 ± 0.71 | 1.95 ± 1.58 | 0.001a |
| RDW (%) | 13.33 ± 1.41 | 13.88 ± 7.81 | 0.165a |
| IBIL (umol/L) | 7.48 ± 7.53 | 11.90 ± 4.73 | < 0.001a |
| Degree of herniation | |||
| Disc herniation | 110 (21.8%) | ||
| Disc prolapse | 388 (77.0%) | ||
| Disc displacement | 1 (0.2%) | ||
| Others | 5 (1%) | ||
| Level of herniation | |||
| L5-S1 | 98 | ||
| L4-L5 and L5-S1 | 160 | ||
| L3-L4 and L4-L5 | 30 | ||
| L4-L5 | 80 | ||
| L3-L4, L4-L5, and L5-S1 | 50 | ||
| L2-L3, L3-L4,L4-L5, and L5-S1 | 12 | ||
| others | 74 |
LDH: Lumbar disc herniation; NEU: Neutrophil ratio; MPV: Mean platelet volume; MON: Mononuclear cell ratio; LYM: Lymphocyte count; RDW: Red blood cell distribution Width; IBIL: Indirect Bilirubin.
ap value was calculated by independent samples t-test. bp value calculated by χ2-test. p < 0.05 indicates statistical significance. The highlighted in bold represent statistical significance.
Association of MIR3142HG polymorphisms and LDH risk
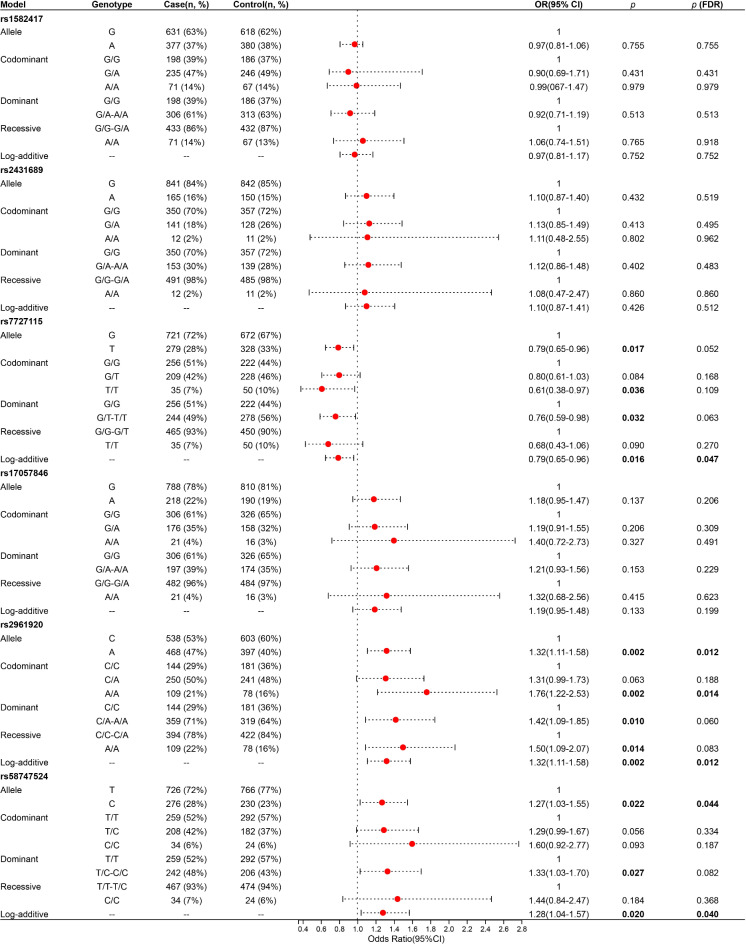
The basic information and allele frequency distribution of the six SNPs in MIR3142HG are shown in Table 2. All SNPs in the control groups conformed to HWE (all p > 0.05). The relationship between MIR3142HG polymorphisms and the risk of LDH was analyzed by logistic regression analysis adjusted for gender and age. As shown in Fig. 1, rs7727115 was associated with a reduced risk of LDH (OR = 0.79, 95%CI = 0.65–0.96, p (FDR) = 0.047). Rs2961920 (OR = 1.76, 95%CI = 1.22–2.53, p (FDR) = 0.014) and rs58747524 (OR = 1.27, 95%CI = 1.03–1.55, p (FDR) = 0.044) showed an increased susceptibility to LDH.
Table 2.
Basic information of the selected SNPs in MIR3142HG.
| SNP ID | Position | Alleles A/B | Callrate | MAF | HWE-p | HaploReg v4.2 | |
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| rs1582417 | 5:159897501 | A/G | 99.9% | 0.374 | 0.381 | 0.343 | Promoter histone marks, Enhancer histone marks, DNAse, Proteins bound, Motifs changed, Selected eQTL hits |
| rs2431689 | 5:159899122 | A/G | 99.5% | 0.164 | 0.151 | 1.000 | Promoter histone marks, Enhancer histone marks, DNAse, Proteins bound, Motifs changed |
| rs7727115 | 5:159901739 | T/G | 99.5% | 0.279 | 0.328 | 0.479 | Promoter histone marks, Enhancer histone marks, DNAse, Motifs changed, Selected eQTL hits |
| rs17057846 | 5:159902313 | A/G | 99.9% | 0.217 | 0.190 | 0.663 | Promoter histone marks, Enhancer histone marks, DNAse, Selected eQTL hits |
| rs2961920 | 5:159911506 | A/C | 99.9% | 0.465 | 0.397 | 0.926 | Enhancer histone marks, Proteins bound, Motifs changed, GRASP eQTL hits |
| rs58747524 | 5:159911584 | C/T | 99.4% | 0.275 | 0.231 | 0.614 | Enhancer histone marks, Proteins bound |
SNP: Single nucleotide polymorphism; HWE: Hardy–Weinberg equilibrium.
p < 0.05 indicates statistical significance.
Fig. 1.
Association of MIR3142HG polymorphisms with LDH susceptibility. SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% confidence interval. p-values were calculated by logistic regression analysis with adjustments for age and gender. p < 0.05 indicates statistical significance.
Stratified analysis by age
We further analyzed the correlation between LDH susceptibility and MIR3142HG polymorphisms under age stratification analysis (Table 3). We observed that rs7727115 had protective effects on the risk of LDH in patients aged ˃ 49 years (OR = 0.71, 95% CI = 0.54–0.93, p (FDR) = 0.025) MIR3142HG polymorphism rs17057846 (OR = 1.59, 95% CI = 1.10–2.30, p (FDR) = 0.029) and rs58747524 (OR = 1.69, 95% CI = 1.18–2.42, p (FDR) = 0.025) were associated with an increased risk of LDH in people aged > 49 years.
Table 3.
The association between SNPs and LDH after stratification by age and gender under different genotypic models.
| SNP ID | Model | Genotype | ≤ 49 years | > 49 years | Male | Female | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p | p (FDR) | OR (95%CI) | p | p (FDR) | OR (95%CI) | p | p (FDR) | OR (95%CI) | p | p (FDR) | |||
| rs1582417 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| A | 1.02 (0.79–1.31) | 0.910 | 0.910 | 0.93 (0.72–1.20) | 0.580 | 0.698 | 1.13 (0.89–1.43) | 0.325 | 0.649 | 0.79 (0.60–1.05) | 0.100 | 0.151 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A | 0.98 (0.67–1.43) | 0.907 | 1.089 | 0.82 (0.56–1.21) | 0.322 | 0.386 | 1.10 (0.78–1.57) | 0.577 | 0.153 | 0.66 (0.43–1.01) | 0.054 | 0.108 | ||
| A/A | 1.06 (0.60–1.86 | 0.837 | 0.837 | 0.96 (0.56–1.65) | 0.876 | 0.876 | 1.30 (0.77–2.20) | 0.320 | 0.640 | 0.69 (0.38–1.24) | 0.214 | 0.321 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A-A/A | 0.99 (0.69–1.43) | 0.977 | 0.977 | 0.85 (0.60–1.23) | 0.391 | 0.469 | 1.14 (0.82–1.59) | 0.428 | 0.642 | 0.67 (0.45–1.00) | 0.047 | 0.071 | ||
| recessive | G/G-G/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.07 (0.64–1.81) | 0.788 | 0.945 | 1.07 (0.65–1.76) | 0.804 | 0.804 | 1.23 (0.76–2.01) | 0.396 | 0.792 | 0.87 (0.51–1.49) | 0.617 | 0.740 | ||
| log-additive | – | 1.02 (0.78–1.32) | 0.909 | 0.909 | 0.94 (0.73–1.21) | 0.630 | 0.756 | 1.13 (0.89–1.44) | 0.318 | 0.636 | 0.79 (0.60–1.05) | 0.099 | 0.148 | |
| rs2431689 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| A | 1.25 (0.89–1.75) | 0.199 | 0.597 | 0.96 (0.68–1.36) | 0.832 | 0.832 | 1.06 (0.78–1.45) | 0.708 | 0.849 | 1.16 (0.80–1.70) | 0.436 | 0.436 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A | 0.15 (0.77–1.70) | 0.497 | 1.490 | 1.12 (0.74–1.68) | 0.593 | 0.593 | 1.02 (0.71–1.47) | 0.898 | 0.898 | 1.30 (0.83–2.03) | 0.254 | 0.304 | ||
| A/A | 2.73 (0.71–10.47) | 0.143 | 0.430 | 0.50 (0.15–1.71) | 0.272 | 0.407 | 1.40 (0.44–4.48) | 0.573 | 0.860 | 0.86 (0.26–2.88) | 0.806 | 0.806 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A-A/A | 1.21 (0.83–1.78) | 0.322 | 0.644 | 1.05 (0.71–1.55) | 0.827 | 0.827 | 1.05 (0.73–1.49) | 0.805 | 0.966 | 1.25 (0.81–1.92) | 0.314 | 0.314 | ||
| recessive | G/G-G/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 2.62 (0.69–10.01) | 0.158 | 0.475 | 0.49 (0.15–1.66) | 0251 | 0.376 | 1.39 (0.44–4.43) | 0.580 | 0.870 | 0.80 (0.24–2.69) | 0.724 | 0.724 | ||
| log-additive | – | 1.26 (0.89–1.78) | 0.192 | 0.577 | 0.97 (0.69–1.37) | 0.874 | 0.874 | 1.06 (0.77–1.46) | 0.706 | 0.847 | 1.16 (0.80–1.68) | 0.443 | 0.443 | |
| rs7727115 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| T | 0.89 (0.68–1.17) | 0.392 | 0.783 | 0.71 (0.54–0.93) | 0.013 | 0.025 | 0.77 (0.60–0.99) | 0.042 | 0.249 | 0.82 (0.61–1.11) | 0.203 | 0.244 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/T | 0.81 (0.56–1.17) | 0.267 | 1.603 | 0.77 (0.53–1.12) | 0.166 | 0.249 | 0.79 (0.56–1.11) | 0.169 | 1.014 | 0.81 (0.54–1.20) | 0.292 | 0.292 | ||
| T/T | 0.89 (0.45–1.77) | 0.746 | 0.895 | 0.44 (0.23–0.84) | 0.014 | 0.081 | 0.57 (0.31–1.03) | 0.064 | 0.383 | 0.68 (0.32–1.44) | 0.311 | 0.373 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/T-T/T | 0.82 (0.58–1.17) | 0.281 | 0.843 | 0.70 (0.49–0.99) | 0.046 | 0.069 | 0.74 (0.54–1.03) | 0.076 | 0.455 | 0.79 (0.53–1.16) | 0.219 | 0.263 | ||
| recessive | G/G-G/T | 1 | 1 | 1 | 1 | |||||||||
| T/T | 0.98 (0.51–1.91) | 0.963 | 0.963 | 0.50 (0.26–0.93) | 0.029 | 0.177 | 0.64 (0.36–1.13) | 0.124 | 0.373 | 0.75 (0.36–1.55) | 0.441 | 0.661 | ||
| log-additive | – | 0.88 (0.67–1.17) | 0.380 | 0.761 | 0.70 (0.54–0.93) | 0.012 | 0.024 | 0.77 (0.60–0.99) | 0.040 | 0.240 | 0.82 (0.60–1.11) | 0.193 | 0.231 | |
| rs17057846 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| A | 0.96 (0.71–1.30) | 0.809 | 0.971 | 1.46 (1.07–2.01) | 0.018 | 0.026 | 1.01 (0.76–1.34) | 0.942 | 0.942 | 1.47 (1.04–2.06) | 0.027 | 0.054 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A | 0.90 (0.62–1.30) | 0.560 | 1.121 | 1.58 (1.08–2.33) | 0.019 | 0.058 | 1.04 (0.74–1.48) | 0.812 | 0.975 | 1.43 (0.95–2.15) | 0.090 | 0.135 | ||
| A/A | 1.18 (0.45–3.08) | 0.731 | 1.096 | 1.63 (0.64–4.17) | 0.307 | 0.368 | 0.93 (0.40–2.16) | 0.861 | 0.861 | 2.92 (0.89–9.55) | 0.077 | 0.154 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A-A/A | 0.92 (0.64–1.32) | 0.645 | 0.967 | 1.59 (1.10–2.30) | 0.014 | 0.029 | 1.03 (0.74–1.44) | 0.863 | 0.863 | 1.51 (1.02–2.26) | 0.042 | 0.085 | ||
| recessive | G/G-G/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.23 (0.48–3.18) | 0.665 | 0.997 | 1.40 (0.55–3.56) | 0.475 | 0.570 | 0.91 (0.40–2.11) | 0.833 | 0.833 | 2.56 (0.79–8.32) | 0.117 | 0.234 | ||
| log-additive | – | 0.96 (0.70–1.31) | 0.798 | 0.957 | 1.46 (1.06–2.01) | 0.020 | 0.030 | 1.01 (0.76–1.35) | 0.940 | 0.940 | 1.51 (1.06–2.15) | 0.022 | 0.044 | |
| rs2961920 | allele | C | 1 | 1 | 1 | 1 | ||||||||
| A | 1.25 (0.97–1.60) | 0.081 | 0.484 | 1.40 (1.09–1.80) | 0.009 | 0.028 | 1.22 (0.97–1.54) | 0.088 | 0.264 | 1.48 (1.12–1.95) | 0.006 | 0.034 | ||
| codominant | C/C | 1 | 1 | 1 | 1 | |||||||||
| C/A | 1.12 (0.75–1.69) | 0.575 | 0.862 | 1.50 (1.02–2.23) | 0.042 | 0.084 | 1.07 (0.74–1.55) | 0.723 | 1.084 | 1.73 (1.11–2.67) | 0.015 | 0.087 | ||
| A/A | 1.62 (0.97–2.71) | 0.065 | 0.393 | 1.87 (1.11–3.14) | 0.018 | 0.055 | 1.53 (0.96–2.43) | 0.073 | 0.218 | 2.15 (1.19–3.88) | 0.011 | 0.067 | ||
| dominant | C/C | 1 | 1 | 1 | 1 | |||||||||
| C/A-A/A | 1.25 (0.85–1.83) | 0.261 | 1.566 | 1.59 (1.10–2.31) | 0.014 | 0.041 | 1.19 (0.84–1.68) | 0.330 | 0.989 | 1.82 (1.20–2.76) | 0.005 | 0.028 | ||
| recessive | C/C–C/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.51 (0.96–2.36) | 0.072 | 0.433 | 1.48 (0.93–2.35) | 0.102 | 0.305 | 1.47 (0.98–2.21) | 0.064 | 0.387 | 1.54 (0.91–2.61) | 0.105 | 0.316 | ||
| log-additive | – | 1.26 (0.97–1.62) | 0.078 | 0.471 | 1.39 (1.08–1.79) | 0.011 | 0.032 | 1.22 (0.97–1.53) | 0.093 | 0.280 | 1.51 (1.13–2.01) | 0.005 | 0.030 | |
| rs58747524 | allele | T | 1 | 1 | 1 | 1 | ||||||||
| C | 1.06 (0.80–1.41) | 0.677 | 1.015 | 1.53 (1.14–2.05) | 0.004 | 0.027 | 1.11 (0.85–1.44) | 0.442 | 0.663 | 1.54 (1.12–2.13) | 0.008 | 0.023 | ||
| codominant | T/T | 1 | 1 | 1 | 1 | |||||||||
| T/C | 1.00 (0.70–1.44) | 0.992 | 0.992 | 1.66 (1.14–2.41) | 0.008 | 0.049 | 1.13 (0.81–1.59) | 0.470 | 1.411 | 1.55 (1.04–2.32) | 0.033 | 0.099 | ||
| C/C | 1.30 (0.61–2.79) | 0.499 | 0.998 | 1.95 (0.88–4.33) | 0.100 | 0.199 | 1.19 (0.60–2.33) | 0.619 | 0.742 | 2.85 (1.06–7.68) | 0.038 | 0.114 | ||
| dominant | T/T | 1 | 1 | 1 | 1 | |||||||||
| T/C–C/C | 1.04 (0.73–1.47) | 0.845 | 1.014 | 1.69 (1.18–2.42) | 0.004 | 0.025 | 1.14 (0.82–1.58) | 0.428 | 0.855 | 1.65 (1.12–2.44) | 0.012 | 0.036 | ||
| recessive | T/T-T/C | 1 | 1 | 1 | 1 | |||||||||
| C/C | 1.30 (0.62–2.74) | 0.491 | 0.981 | 1.60 (0.73–3.49) | 0.242 | 0.484 | 1.13 (0.58–2.18) | 0.724 | 0.869 | 2.38 (0.90–6.32) | 0.082 | 0.491 | ||
| log-additive | – | 1.07 (0.80–1.42) | 0.667 | 1.000 | 1.53 (1.14–2.07) | 0.005 | 0.031 | 1.11 (0.85–1.45) | 0.434 | 0.651 | 1.60 (1.15–2.24) | 0.006 | 0.018 | |
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% confidence interval; FDR: False discovery rate.
p < 0.05 indicates SNP with statistical significance. The highlighted in bold represent significant association.
Stratified analysis by gender
As shown in Table 3, gender-based stratification analysis found that rs17057846, rs2961920, and rs58747524 in MIR3142HG were associated with an increased risk of LDH in women. Specifically, rs17057846 had a higher risk probability to LDH (OR = 1.51, 95% CI = 1.06–2.15, p (FDR) = 0.044). We also found rs2961920 (OR = 1.82, 95% CI = 1.20–2.76, p (FDR) = 0.028) and rs58747524 (OR = 1.54, 95% CI = 1.12–2.13, p (FDR) = 0.023) had an increased risk of LDH.
Stratified analysis by severity of LDH
When stratified by the severity of LDH (Table 4), we found that rs7727115 was linked with decreased susceptibility to LDH among disc prolapse individuals (OR = 0.79, 95%CI = 0.64–0.97, p (FDR) = 0.044). Besides, rs2961920 (OR = 1.86, 95%CI = 1.26–2.74, p (FDR) = 0.010) and rs58747524 (OR = 1.32, 95%CI = 1.05–1.65, p (FDR) = 0.047) were related to an increased risk of LDH in patients with disc prolapse.
Table 4.
The association between SNPs and LDH stratified by severity of lumbar disc herniation and hernia levels.
| SNP ID | Model | Genotype | Disc herniation | Disc prolapse | Single hernia | Multiple hernias | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p | p (FDR) | OR (95%CI) | p | p (FDR) | OR (95%CI) | p | p (FDR) | OR (95%CI) | p | p (FDR) | |||
| rs1582417 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| A | 1.04 (0.77–1.41) | 0.779 | 0.779 | 0.96 (0.79–1.17) | 0.678 | 0.678 | 1.05 (0.82–1.35) | 0.677 | 0.677 | 0.94 (0.76–1.18) | 0.605 | 0.726 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A | 0.97 (0.62–1.52) | 0.886 | 0.886 | 0.88 (0.65–1.18) | 0.394 | 0.394 | 0.85 (0.58–1.24) | 0.385 | 0.577 | 0.98 (0.71–1.36) | 0.916 | 0.916 | ||
| A/A | 1.17 (0.62–2.20) | 0.626 | 0.939 | 0.97 (0.64–1.47) | 0.888 | 0.888 | 1.26 (0.75–2.10) | 0.383 | 0.460 | 0.85 (0.52–1.40) | 0.531 | 0.637 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A-A/A | 1.01 (0.66–1.55) | 0.962 | 0.962 | 0.90 (0.69–1.18) | 0.455 | 0.455 | 0.93 (0.65–1.33) | 0.698 | 0.837 | 0.95 (0.70–1.31) | 0.771 | 0.771 | ||
| recessive | G/G-G/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.19 (0.67–2.13) | 0.551 | 0.827 | 1.04 (0.71–1.53) | 0.841 | 1.009 | 1.38 (0.86–2.20) | 0.181 | 0.542 | 0.86 (0.54–1.37) | 0.527 | 0.632 | ||
| log-additive | – | 1.05 (0.78–1.43) | 0.735 | 0.882 | 0.96 (0.79–1.17) | 0.664 | 0.664 | 1.06 (0.82–1.36) | 0.677 | 0.677 | 0.94 (0.75–1.18) | 0.596 | 0.715 | |
| rs2431689 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| A | 1.06 (0.71–1.59) | 0.769 | 0.922 | 1.12 (0.87–1.45) | 0.377 | 0.452 | 1.35 (0.98–1.85) | 0.062 | 0.123 | 0.95 (0.70–1.29) | 0.744 | 0.744 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A | 1.04 (0.65–1.67) | 0.874 | 1.048 | 1.16 (0.86–1.57) | 0.324 | 0.389 | 1.50 (1.03–2.18) | 0.034 | 0.207 | 0.89 (0.63–1.28) | 0.541 | 0.649 | ||
| A/A | 1.27 (0.34–4.67) | 0.720 | 0.864 | 1.09 (0.45–2.68) | 0.845 | 1.014 | 1.19 (0.37–3.85) | 0.771 | 0.771 | 1.17 (0.44–3.08) | 0.751 | 0.751 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A-A/A | 1.06 (0.67–1.67) | 0.811 | 0.973 | 1.16 (0.86–1.55) | 0.326 | 0.391 | 1.48 (1.02–2.13) | 0.038 | 0.113 | 0.92 (0.65–1.29) | 0.623 | 0.747 | ||
| recessive | G/G-G/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.26 (0.34–4.60) | 0.730 | 0.876 | 1.05 (0.43–2.56) | 0.917 | 0.917 | 1.05 (0.33–3.39) | 0.929 | 0.929 | 1.20 (0.46–3.15) | 0.708 | 0.708 | ||
| log-additive | – | 1.07 (0.71–1.59) | 0.754 | 0.754 | 1.13 (0.87–1.46) | 0.366 | 0.439 | 1.37 (0.99–1.89) | 0.059 | 0.119 | 0.95 (0.70–1.29) | 0.750 | 0.750 | |
| rs7727115 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| T | 0.80 (0.58–1.11) | 0.184 | 1.102 | 0.79 (0.64–0.97) | 0.022 | 0.044 | 0.75 (0.57–0.99) | 0.039 | 0.117 | 0.78 (0.61–0.98) | 0.036 | 0.071 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/T | 0.80 (0.52–1.23) | 0.308 | 0.923 | 0.80 (0.60–1.05) | 0.108 | 0.216 | 0.75 (0.52–1.08) | 0.122 | 0.244 | 0.80 (0.58–1.10) | 0.162 | 0.323 | ||
| T/T | 0.63 (0.28–1.40) | 0.254 | 0.762 | 0.59 (0.35–0.98) | 0.042 | 0.126 | 0.56 (0.28–1.12) | 0.101 | 0.302 | 0.54 (0.29–0.98) | 0.043 | 0.130 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/T-T/T | 0.77 (0.51–1.16) | 0.210 | 0.632 | 0.76 (0.58–0.99) | 0.042 | 0.085 | 0.72 (0.51–1.02) | 0.061 | 0.122 | 0.75 (0.55–1.02) | 0.064 | 0.128 | ||
| recessive | G/G-G/T | 1 | 1 | 1 | 1 | |||||||||
| T/T | 0.70 (0.32–1.52) | 0.365 | 1.095 | 0.66 (0.40–1.08) | 0.094 | 0.282 | 0.64 (0.32–1.26) | 0.193 | 0.290 | 0.60 (0.33–1.07) | 0.085 | 0.510 | ||
| log-additive | – | 0.79 (0.57–1.10) | 0.170 | 1.020 | 0.78 (0.63–0.96) | 0.020 | 0.041 | 0.75 (0.57–0.99) | 0.042 | 0.127 | 0.76 (0.60–0.97) | 0.028 | 0.083 | |
| rs17057846 | allele | G | 1 | 1 | 1 | 1 | ||||||||
| A | 1.19 (0.83–1.70) | 0.340 | 0.509 | 1.18 (0.94–1.49) | 0.159 | 0.239 | 1.09 (0.80–1.47) | 0.584 | 0.700 | 1.25 (0.96–1.62) | 0.099 | 0.149 | ||
| codominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A | 1.25 (0.81–1.93) | 0.320 | 0.640 | 1.17 (0.88–1.56) | 0.280 | 0.420 | 1.02 (0.70–1.48) | 0.923 | 0.923 | 1.29 (0.93–1.78) | 0.122 | 0.365 | ||
| A/A | 1.20 (0.39–3.72) | 0.751 | 0.751 | 1.47 (0.73–2.97) | 0.284 | 0.426 | 1.50 (0.62–3.62) | 0.369 | 0.554 | 1.48 (0.67–3.29) | 0.331 | 0.496 | ||
| dominant | G/G | 1 | 1 | 1 | 1 | |||||||||
| G/A-A/A | 1.24 (0.81–1.90) | 0.314 | 0.627 | 1.20 (0.91–1.58) | 0.199 | 0.298 | 1.06 (0.74–1.52) | 0.745 | 0.745 | 1.31 (0.96–1.79) | 0.091 | 0.137 | ||
| recessive | G/G-G/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.11 (0.36–3.40) | 0.855 | 0.855 | 1.39 (0.69–2.79) | 0.352 | 0.704 | 1.49 (0.62–3.57) | 0.372 | 0.446 | 1.36 (0.62–2.98) | 0.447 | 0.671 | ||
| log-additive | – | 1.19 (0.83–1.71) | 0.350 | 0.700 | 1.19 (0.94–1.05) | 0.157 | 0.235 | 1.10 (0.80–1.49) | 0.563 | 0.676 | 1.26 (0.97–1.65) | 0.088 | 0.132 | |
| rs2961920 | allele | C | 1 | 1 | 1 | 1 | ||||||||
| A | 1.20 (0.89–1.61) | 0.230 | 0.689 | 1.36 (1.12–1.64) | 0.002 | 0.010 | 1.44 (1.13–1.83) | 0.004 | 0.022 | 1.27 (1.02–1.57) | 0.033 | 0.099 | ||
| codominant | C/C | 1 | 1 | 1 | 1 | |||||||||
| C/A | 1.31 (0.82–2.10) | 0.263 | 1.580 | 1.32 (0.97–1.79) | 0.075 | 0.226 | 1.49 (0.99–2.25) | 0.054 | 0.163 | 1.28 (0.91–1.81) | 0.161 | 0.323 | ||
| A/A | 1.40 (0.76–2.59) | 0.286 | 0.571 | 1.86 (1.26–2.74) | 0.002 | 0.010 | 2.07 (1.25–3.43) | 0.005 | 0.030 | 1.63 (1.04–2.53) | 0.032 | 0.193 | ||
| dominant | C/C | 1 | 1 | 1 | 1 | |||||||||
| C/A-A/A | 1.33 (0.85–2.08) | 0.211 | 0.632 | 1.45 (1.09–1.94) | 0.011 | 0.033 | 1.63 (1.11–2.40) | 0.013 | 0.080 | 1.37 (0.99–1.90) | 0.061 | 0.184 | ||
| recessive | C/C–C/A | 1 | 1 | 1 | 1 | |||||||||
| A/A | 1.19 (0.69–2.05) | 0.528 | 1.057 | 1.58 (1.12–2.21) | 0.009 | 0.053 | 1.61 (1.05–2.48) | 0.031 | 0.184 | 1.40 (0.95–2.08) | 0.090 | 0.271 | ||
| log-additive | – | 1.20 (0.89–1.62) | 0.230 | 0.689 | 1.36 (1.12–1.64) | 0.002 | 0.010 | 1.44 (1.12–1.85) | 0.004 | 0.025 | 1.03 (1.03–1.59) | 0.029 | 0.057 | |
| rs58747524 | allele | T | 1 | 1 | 1 | 1 | ||||||||
| C | 1.18 (0.84–1.65) | 0.336 | 0.673 | 1.29 (1.04–1.61) | 0.019 | 0.057 | 1.21 (0.92–1.60) | 0.173 | 0.259 | 1.36 (1.07–1.73) | 0.013 | 0.079 | ||
| codominant | T/T | 1 | 1 | 1 | 1 | |||||||||
| T/C | 0.96 (0.61–1.50) | 0.842 | 1.263 | 1.41 (1.07–1.86) | 0.014 | 0.087 | 1.14 (0.79–1.64) | 0.480 | 0.576 | 1.49 (1.08–2.05) | 0.014 | 0.084 | ||
| C/C | 1.93 (0.87–4.25) | 0.104 | 0.623 | 1.46 (0.80–2.65) | 0.221 | 0.441 | 1.70 (0.82–3.49) | 0.152 | 0.303 | 1.69 (0.87–3.27) | 0.119 | 0.238 | ||
| dominant | T/T | 1 | 1 | 1 | 1 | |||||||||
| T/C–C/C | 1.07 (0.70–1.63) | 0.756 | 1.134 | 1.42 (1.09–1.85) | 0.011 | 0.063 | 1.20 (0.85–1.71) | 0.296 | 0.445 | 1.51 (1.11–2.05) | 0.008 | 0.048 | ||
| recessive | T/T-T/C | 1 | 1 | 1 | 1 | |||||||||
| C/C | 1.96 (0.91–4.24) | 0.087 | 0.523 | 1.26 (0.70–2.26) | 0.447 | 0.671 | 1.61 (0.79–3.26) | 0.187 | 0.374 | 1.42 (0.75–2.71) | 0.283 | 0.567 | ||
| log-additive | – | 1.17 (0.84–1.64) | 0.357 | 0.535 | 1.32 (1.05–1.65) | 0.016 | 0.047 | 1.22 (0.92–1.62) | 0.172 | 0.259 | 1.40 (1.09–1.79) | 0.009 | 0.057 | |
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% confidence interval; FDR: False discovery rate.
p < 0.05 indicates SNP with statistical significance. The highlighted in bold represent significant association.
Stratified by hernia levels
As demonstrated in Table 4, it was found that rs2431689 was significantly associated with increased susceptibility to patients with single hernia (OR = 1.50, 95%CI = 1.03–2.18, p (FDR) = 0.207). Besides, rs58747524 demonstrated an increased risk correlation with LDH within multiple hernias (OR = 1.51, 95%CI = 1.11–2.05, p (FDR) = 0.048).
SNP–SNP interaction analysis using GMDR and MDR
GMDR analysis (Table 5) showed that the six-locus model with a combination of rs1582417, rs2431689, rs7727115, rs17057846, rs2961920, and rs58747524 was the best predictive model for LDH (test balanced accuracy = 0.5149, CVC = 10/10). Additionally, the six-locus model was associated with increased susceptibility to LDH (OR = 1.8899, 95%CI = 1.3227–2.7003, p = 0.0004). We further used the information gain theory to explain the relationship between six SNPs and drew a tree diagram. As shown in Fig. 2, the interaction map with low entropy values or negative entropy values (plotted in orange, blue, or green) indicated the independence or redundancy of each pairwise combination of attributes.
Table 5.
SNP–SNP interaction models of the MIR3142HG gene analyzed by the GMDRa.
| Model | Training Bal. Acc | Testing Bal. Acc.b | CVC |
|---|---|---|---|
| rs2961920 | 0.5391 | 0.5159 | 6/10 |
| rs2431689, rs58747524 | 0.5487 | 0.5145 | 7/10 |
| rs2431689, rs2961920, rs58747524 | 0.5568 | 0.5034 | 5/10 |
| rs1582417,rs7727115, rs2961920, rs58747524 | 0.5660 | 0.4880 | 4/10 |
| rs1582417, rs7727115, rs17057846, rs2961920, rs58747524 | 0.5754 | 0.5036 | 5/10 |
| rs1582417, rs2431689, rs7727115, rs17057846, rs2961920, rs58747524 | 0.5802 | 0.5149 | 10/10 |
a Whole dataset statistics: Training Balanced Accuracy, 0.5782; Training Accuracy, 0.5782; Training Sensitivity, 0.5302; Training Specificity, 0.6261; Training Odds Ratio, 1.8899 (1.3227, 2.7003); Training χ2 (P), 12.3300 (p = 0.0004); Training Precision, 0.5860; Training Kappa, 0.1563; Training F-Measure, 0.5567.
GMDR: Generalized Multifactor dimensionality reduction; Bal. Acc: Balanced accuracy; CVC: Cross‐validation consistency.
Fig. 2.
The tree diagram analysis of SNP interaction. Values in nodes represent the information gains of individual attribute (main effects). Values between nodes are information gains of each pair of attributes (interaction effects). The brown with positive percent entropy represents a moderate level of synergy on the phenotype. The green and blue with negative values indicate redundancy or lack of synergy between the SNPs.
The relationship between SNP genotypes and clinical indicators in LDH
By analyzing the relationship between SNP genotypes and clinical parameters in LDH (Table 6), we found that rs1582417 and rs7727115 different genotypes had significantly different IBIL levels (p = 0.026, p = 0.048, respectively). Similarly, the genotypes of rs17057846, rs2961920, and rs58747524 in LDH patients showed significantly different MON levels (p = 0.004, p = 0.026, and p = 0.021, respectively). Rs7727115 genotypes were significantly associated with RDW levels (p = 0.007). Besides, AA (2.094 ± 0.300 109/L) and AG genotypes (1.719 ± 0.069 109/L) in rs2431689 were related to an increased concentration of LYM in LDH patients compared with GG genotype (0.624 ± 0.035 109/L) (p = 0.042).
Table 6.
The relationship between genotypes at different loci and clinical parameters.
| rs1582417 | rs2431689 | rs7727115 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | AA | GA | GG | p | AA | AG | GG | p | TT | GT | GG | p |
| NEU | 67.688 ± 1.729 | 68.121 ± 1.050 | 70.326 ± 0.940 | 0.229 | 66.940 ± 3.402 | 67.655 ± 1.405 | 69.484 ± 0.756 | 0.419 | 71.469 ± 1.937 | 68.780 ± 1.078 | 68.593 ± 0.915 | 0.564 |
| MPV | 11.594 ± 0.141 | 11.477 ± 0.095 | 11.398 ± 0.094 | 0.568 | 11.483 ± 0.332 | 11.506 ± 0.106 | 11.443 ± 0.076 | 0.897 | 11.153 ± 0.221 | 11.408 ± 0.104 | 11.553 ± 0.077 | 0.201 |
| MON | 6.884 ± 0.275 | 6.788 ± 0.151 | 6.506 ± 0.169 | 0.343 | 6.403 ± 0.422 | 6.829 ± 0.197 | 6.647 ± 0.127 | 0.673 | 6.464 ± 0.320 | 6.724 ± 0.172 | 6.668 ± 0.142 | 0.830 |
| LYM | 1.810 ± 0.108 | 1.653 ± 0.044 | 1.615 ± 0.047 | 0.138 | 2.094 ± 0.300 | 1.719 ± 0.069 | 0.624 ± 0.035 | 0.042 | 1.604 ± 0.093 | 1.643 ± 0.051 | 1.686 ± 0.045 | 0.718 |
| RDW | 13.352 ± 0.148 | 13.358 ± 0.106 | 13.291 ± 0.087 | 0.880 | 13.583 ± 0.358 | 13.338 ± 0.135 | 13.320 ± 0.072 | 0.817 | 13.147 ± 0.159 | 13.575 ± 0.121 | 13.173 ± 0.070 | 0.007 |
| IBIL | 9.800 ± 1.958 | 6.991 ± 0.318 | 7.262 ± 0.399 | 0.026 | 6.908 ± 0.725 | 7.482 ± 0.381 | 7.485 ± 0.467 | 0.967 | 5.821 ± 0.405 | 6.800 ± 0.267 | 8.299 ± 0.642 | 0.048 |
| rs17057846 | rs2961920 | rs58747524 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | AA | GA | GG | p | AA | CA | CC | p | TT | CT | CC | p |
| NEU | 75.164 ± 2.854 | 69.032 ± 1.067 | 68.391 ± 0.873 | 0.125 | 68.873 ± 1.495 | 68.646 ± 0.914 | 69.583 ± 1.245 | 0.831 | 68.510 ± 0.951 | 68.892 ± 1.014 | 71.639 ± 2.121 | 0.520 |
| MPV | 11.457 ± 0.215 | 11.508 ± 0.967 | 11.443 ± 0.080 | 0.880 | 11.422 ± 0.118 | 11.567 ± 0.080 | 11.321 ± 0.132 | 0.214 | 11.408 ± 0.091 | 11.544 ± 0.086 | 11.309 ± 0.229 | 0.457 |
| MON | 5.033 ± 0.444 | 6.789 ± 0.183 | 6.740 ± 0.130 | 0.004 | 6.242 ± 0.222 | 6.943 ± 0.144 | 6.575 ± 0.203 | 0.026 | 6.734 ± 0.143 | 6.802 ± 0.166 | 5.607 ± 0.367 | 0.021 |
| LYM | 1.375 ± 0.131 | 1.639 ± 0.048 | 1.696 ± 0.043 | 0.116 | 1.684 ± 0.070 | 1.686 ± 0.047 | 1.594 ± 0.054 | 0.431 | 1.682 ± 0.481 | 1.654 ± 0.045 | 1.547 ± 0.116 | 0.581 |
| RDW | 13.143 ± 0.270 | 13.206 ± 0.090 | 13.421 ± 0.088 | 0.228 | 13.159 ± 0.118 | 13.325 ± 0.087 | 13.479 ± 0.133 | 0.208 | 44.502 ± 0.375 | 43.961 ± 0.244 | 44.000 ± 0.648 | 0.125 |
| IBIL | 10.395 ± 2.851 | 7.317 ± 0.415 | 7.367 ± 0.474 | 0.209 | 8.032 ± 0.662 | 7.240 ± 0.317 | 7.481 ± 0.939 | 0.673 | 7.392 ± 0.551 | 7.327 ± 0.371 | 9.175 ± 1.811 | 0.423 |
NEU: Neutrophil ratio; MPV: Mean platelet volume; MON: Mononuclear cell ratio; LYM: Lymphocyte count; RDW: Red blood cell distribution Width; IBIL: Indirect Bilirubin.
p < 0.05 indicates statistical significance. The highlighted in bold represent statistical significance.
Impact of SNPs on gene expression (eQTLs)
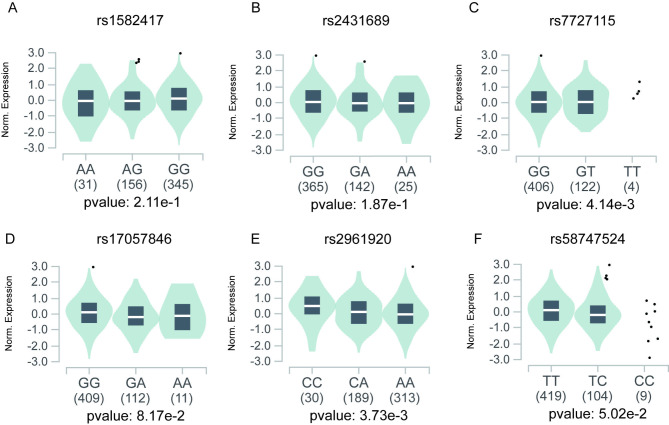
We further evaluated each SNP’s expression loci (eQTLs) by GTEx. As shown in Fig. 3, we found that rs7727115 (Fig. 3C) and rs2961920 (Fig. 3E) were significantly associated with mRNA expression of MIR3142HG. Whereas there were no significant associations between rs1582417, rs2431689, rs17057846, rs58747524 and MIR3142HG expression (Fig. 3A, B, D, F).
Fig. 3.
Functional relevance of each SNP on gene expression of MIR3142HG in GTEx database. (A) rs1582417, (B) rs2431689, (C) rs7727115, (D) rs17057846, (E) rs2961920, and (F) rs58747524.
Discussion
LDH is a degenerative disease that can lead to neuropathic symptoms such as spinal cord pain syndrome and nerve root ischemia 23. Evidence has shown that inflammation-related factors play an important role in LDH, can accelerate inflammation and disc formation, and ultimately deepen lumbar disc degeneration and pain 24. MIR3142HG, also known as the MIR3142 host gene, is located on human chromosome 5q33.3. Studies showed that MIR3142HG is abnormally expressed in lung fibroblasts induced by interleukin 1β (IL-1β), which plays an important role in inflammation regulation 14. Studies have reported that SNP can significantly influence gene expression 25,26. We speculate that MIR3142HG polymorphisms have some possible roles in the occurrence of LDH. In this study, we explored the relationship between the MIR3142HG polymorphism and LDH susceptibility in the Chinese Han population. We found that individuals carrying the T and TT genotype of rs7727115 in MIR3142HG had a reduced risk of developing LDH compared to the other genotypes, whereas rs17057846 (G > A), rs2961920 (C > A), and rs58747524 (T > C) were associated with an increased susceptibility to LDH. To the best of our knowledge, this is the first study of MIR3142HG polymorphism in LDH, which can serve as potential genetic markers for LDH risk assessment, which may contribute to early diagnosis, personalized treatment strategies, and preventive measures for LDH patients.
Our study indicated that rs7727115 was associated with a reduced risk of LDH, and rs17057846, rs2961920, and rs58747524 were associated with an increased susceptibility to LDH. According to the present results, a previous study has demonstrated that rs17057846, rs2961920, and rs58747524 are significantly associated with an increased risk of glioma 27. Besides, rs7727115 could reduce the risk of steroid-induced osteonecrosis of the femoral head 20. These findings suggest that MIR3142HG polymorphisms have a crucial role in the progression of human diseases including LDH. Several studies have provided more and more evidence that intron SNPs can confer human diseases susceptibility by affecting gene expression 28–30. Here, we evaluated the impact of SNPs on the mRNA expression of MIR3142HG, we found that rs17057846 and rs2961920 were significantly associated with MIR3142HG expression. Besides, the possible functions of MIR3142HG SNPs were predicted and we found that rs17057846 could influence Promoter histone marks, Enhancer histone marks, DNAse, and Selected eQTL hits. Rs2961920 might be involved in Enhancer histone marks, Proteins bound, Motifs changed, and GRASP eQTL hits. Taken above, we speculated that MIR3142HG SNPs, especially rs17057846 and rs2961920 may affect gene expression and function, and then contribute to LDH development, which needs further study and confirmation.
Previously published studies have revealed that age is the risk factor for LDH, and the number of individuals diagnosed with symptomatic cervical and lumbar disc herniation increased with age 31–33. In this study, the mean age was 49 years in the case and control groups, thus we stratified by 49 years. Age-stratified analysis showed that rs7727115 reduced the risk of LDH in patients aged > 49 years, and rs17057846, rs2961920, and rs58747524 had a risk-increasing influence on the patients aged > 49 years, but not in aged ≤ 49 years. Similar to our results, rs9450607 in the Eyes Shut Homolog (EYS) gene was related to an increased risk of LDH in people aged ≥ 49 years, but not in those aged < 49 years 34. Additionally, rs6265, rs11030104, and rs10767664 in Brain-Derived Neurotrophic Factor (BDNF) could significantly increase the risk of LDH in those aged > 50 years, but not in those aged ≤ 50 years 35. Rs77681114 polymorphism in Gasdermin C (GSDMC) protected LDH risk at age ≥ 49 years, but not at age < 49 years 36. To sum up, these findings indicated that the association between genetic polymorphisms and LDH susceptibility may depend on age. Gender is also another risk factor for LDH, and the incidence of LDH was higher in women than in men 32. When stratified by gender, we observed that rs17057846, rs2961920, and rs58747524 had a risk-increasing influence on LDH in females, but not in males. In consistent with our results, Zhu et al. found that Interleukin 1 Receptor Type 1 (IL1R1) rs956730 was statistically significant in LDH among males, but not in females 24. The MMP-3 gene rs591058 was related to an increased susceptibility to LDH in females, but not in males 37. Besides, rs1008993 was associated with increased LDH risk in males, but not in females 38. These results suggest that genetic susceptibility to LDH is influenced by gender. Besides, we found that rs7727115, rs2961920, rs2431689, and rs58747524 also were associated with the risk of severity of LDH and hernia levels. Taken above, our study given that the association of MIR3142HG polymorphisms and LDH risk may rely on age, gender, severity of LDH, and hernia levels, highlights the importance of considering heterogeneity in studies of the association between genetics and LDH.
MPV and RDW are important indicators for evaluating the inflammatory state and immune response of the body. Dagistan et al. found that MPV and RDW may be related to the occurrence of LDH 5. Our study showed that rs7727115 genotypes were significantly associated with RDW levels. Besides, AA and AG genotypes in rs2431689 were related to an increased concentration of LYM in LDH patients. We speculate that certain genotypes (such as rs 7727115 and rs 2431689) may influence the levels of these indicators, thereby affecting the immune response in LDH, which needed to be further validated.
Inevitably, the current study has some limitations. First, the present study focuses on the Chinese Han population, the association is needed to be verified in another ethnicity. Second, some potential risk factors, such as activity status and occupation, were not included in our analysis due to the limited information, which should be further evaluated in future studies. Third, our study is the basic research, the association between MIR3142HG polymorphisms and the expression of MIR3142HG need to be further investigated. Despite the above limitations, this is the first study to determine the impact of MIR3142HG polymorphisms on LDH risk.
Conclusion
This study indicated that rs2431689, rs7727115, rs17057846, rs2961920, and rs58747524 in MIR3142HG were significantly associated with LDH susceptibility in the Chinese population, which may serve as a new biomarker for prevention and diagnosis of LDH.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank all participants in this study for providing blood samples.
Author contributions
Qi Dong contributed to methodology, formal analysis, software, validation, and writing. Guoxia Ren contributed to methodology, formal analysis, and interpretation. Dingjun Hao contributed to conceiving the study and revising the manuscript. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University. The participants provided their written informed consent to participate in this study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
They are co-first authors.
References
- 1.Han, L. et al. Short-term study on risk-benefit outcomes of two spinal manipulative therapies in the treatment of acute radiculopathy caused by lumbar disc herniation: study protocol for a randomized controlled trial. Trials16, 122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, H., Yu, H., Ren, D., Zhu, K. & Huang, H. Plumbagin exerts protective effects in nucleus pulposus cells by attenuating hydrogen peroxide-induced oxidative stress, inflammation and apoptosis through NF-κB and Nrf-2. Int. J. Mol. Med.37(6), 1669–1676 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Rogerson, A., Aidlen, J. & Jenis, L. G. Persistent radiculopathy after surgical treatment for lumbar disc herniation: causes and treatment options. Int. Orthopaed.43(4), 969–973 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Teichtahl, A. J. et al. A Dose-response relationship between severity of disc degeneration and intervertebral disc height in the lumbosacral spine. Arthritis Res. Therapy17, 297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagistan, Y. et al. Could red cell distribution width and mean platelet volume be a predictor for lumbar disc hernias?. Ideggyogyaszati Szemle69(11–12), 411–414 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Bozkurt, H., Arac, D. & Cigdem, B. Effect of preoperative uric acid level and neutrophil/lymphocyte ratio on preoperative and postoperative visual analogue pain scores in patients with lumbar disc herniation: A cross-sectional study. Turk. Neurosurg.29(5), 705–709 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Battié, M. C., Videman, T., Levälahti, E., Gill, K. & Kaprio, J. Genetic and environmental effects on disc degeneration by phenotype and spinal level: a multivariate twin study. Spine33(25), 2801–2808 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Zhu, Y. et al. Association between IL4, IL6 gene polymorphism and lumbar disc degeneration in Chinese population. Oncotarget8(51), 89064–89071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer, J. E. et al. Genetic polymorphisms associated with intervertebral disc degeneration. Spine J.13(3), 299–317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, X. et al. Investigation of the STOX1 polymorphism on lumbar disc herniation. Mol. Genet. Genomic Med.8(1), e1038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, W. K. et al. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Proliferation50(1), e12313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. et al. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulposus cells via regulating Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun.491(3), 668–674 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Mi, D. et al. Long non-coding RNA FAF1 promotes intervertebral disc degeneration by targeting the Erk signaling pathway. Mol. Med. Rep.17(2), 3158–3163 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Hadjicharalambous, M. R. et al. Long Non-coding RNAs Are Central Regulators of the IL-1β-Induced Inflammatory Response in Normal and Idiopathic Pulmonary Lung Fibroblasts. Front. Immunol.9, 2906 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, Y., Li, S., Dong, R. & Li, X. Long noncoding RNA MIR3142HG accelerates lipopolysaccharide-induced acute lung injury via miR-95-5p/JAK2 axis. Human Cell35(3), 856–870 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Liu, C., Fei, H. D., Sun, Z. Y. & Tian, J. W. Bioinformatic analysis of the microarray gene expression profile in degenerative intervertebral disc cells exposed to TNF-α. Eur. Rev. Med. Pharmacol. Sci.19(18), 3332–3339 (2015). [PubMed] [Google Scholar]
- 17.Zhou, T. et al. Mechanism of microRNA-146a-mediated IL-6/STAT3 signaling in lumbar intervertebral disc degeneration. Exp. Therapeutic Med.14(2), 1131–1135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, F. et al. Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer118(4), 937–946 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Cao Y, Wang R, Zhang H, Zhai P, Wei J. Genetic variants in MIR3142HG contribute to the predisposition of IgA nephropathy in a Chinese Han population. Public Health Genomics 1–11 (2022). [DOI] [PubMed]
- 20.Wang T, Wu H, Sun M, Liu T, An F, Dong Q, et al. Association between genetic polymorphisms of MIR3142HG and the risk of steroid-induced osteonecrosis of the femoral head in the population of Northern China. Public Health Genomics 1–9 (2021). [DOI] [PubMed]
- 21.Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res.50(D1), D988–D995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie, M. D., Hahn, L. W. & Moore, J. H. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genetic Epidemiol.24(2), 150–157 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Liu, Q., Jin, L., Shen, F. H., Balian, G. & Li, X. J. Fullerol nanoparticles suppress inflammatory response and adipogenesis of vertebral bone marrow stromal cells–a potential novel treatment for intervertebral disc degeneration. Spine J.13(11), 1571–1580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, Y. et al. IL1R1 polymorphisms are associated with lumbar disc herniation risk in the Northwestern Chinese Han Population. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res.25, 3728–3738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iurescia, S., Seripa, D. & Rinaldi, M. Looking beyond the 5-HTTLPR polymorphism: Genetic and epigenetic layers of regulation affecting the serotonin transporter gene expression. Mol. Neurobiol.54(10), 8386–8403 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Horiuchi, Y. et al. A polymorphism in the PDLIM5 gene associated with gene expression and schizophrenia. Biol. Psychiatry59(5), 434–439 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Guo, X. et al. Evaluation of genetic variants in MIR3142HG in susceptibility to and prognosis of glioma. Am. J. Clin. Oncol.43(1), 1–8 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Zhao, H. et al. An intronic variant associated with systemic lupus erythematosus changes the binding affinity of Yinyang1 to downregulate WDFY4. Genes Immunity13(7), 536–542 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Seo, S. et al. Functional analysis of deep intronic SNP rs13438494 in intron 24 of PCLO gene. PLoS ONE8(10), e76960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, D. & Sadee, W. CYP3A4 intronic SNP rs35599367 (CYP3A4*22) alters RNA splicing. Pharmacogenet. Genom.26(1), 40–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karjalainen, U. et al. Role of environmental factors and history of low back pain in sciatica symptoms among Finnish adolescents. Spine38(13), 1105–1111 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Kim YK, Kang D, Lee I, Kim SY. Differences in the incidence of symptomatic cervical and lumbar disc herniation according to age, sex and national health insurance eligibility: A pilot study on the disease’s association with work. Int. J. Environ. Res. Public Health 15(10) (2018). [DOI] [PMC free article] [PubMed]
- 33.Zhang, Y. G. et al. A controlled case study of the relationship between environmental risk factors and apoptotic gene polymorphism and lumbar disc herniation. Am. J. Pathol.182(1), 56–63 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Ji, D. et al. Correlation of EYS polymorphisms with lumbar disc herniation risk among Han Chinese population. Mol. Genet. Genomic Med.7(9), e890 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, Y. et al. Associations between variants in BDNF/BDNFOS gene and lumbar disc herniation risk among han Chinese people. Sci. Rep.8(1), 12782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, J. et al. Association of GSDMC polymorphisms with lumbar disc herniation among Chinese Han population. Int. J. Immunogenet.47(6), 546–553 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Luo, Y. et al. Interactions between the MMP-3 gene rs591058 polymorphism and occupational risk factors contribute to the increased risk for lumbar disk herniation: A case-control study. J. Clin. Lab. Anal.34(7), e23273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu, B. et al. Association of glypican-6 polymorphisms with lumbar disk herniation risk in the Han Chinese population. Mol. Genet. Genomic Med.7(7), e00747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.