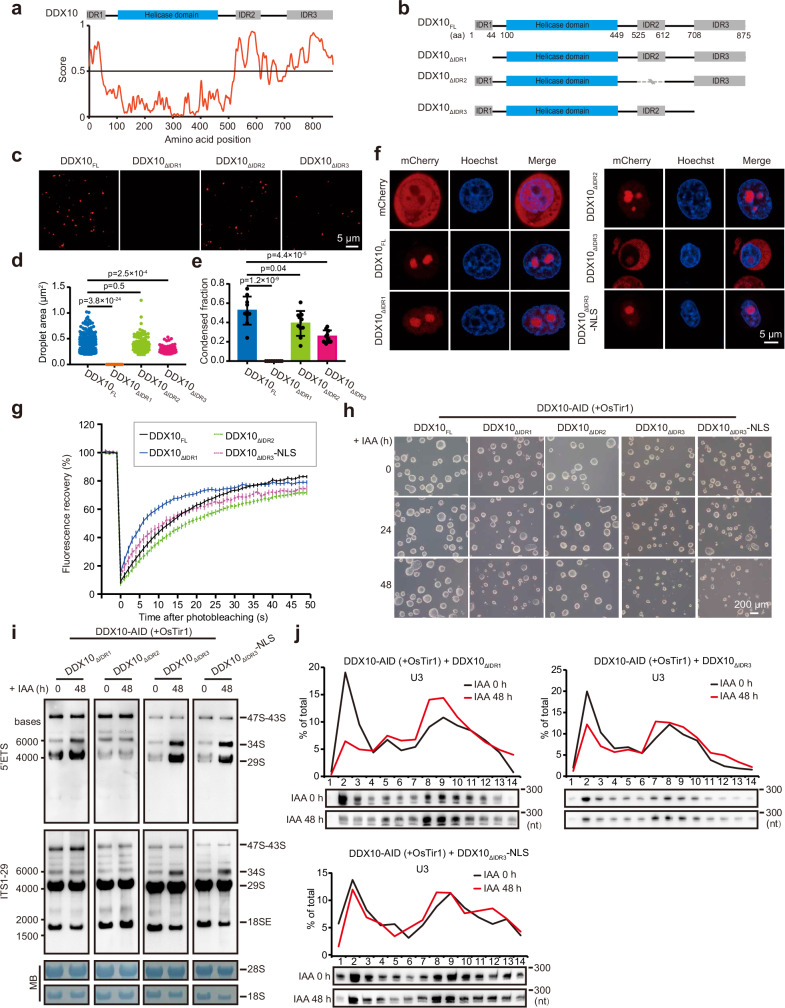
Fig. 5. DDX10 drives LLPS to modulate ribosome biogenesis.
a Top: Domain mapping of mouse DDX10 displaying a helicase domain and three intrinsically disordered regions (IDR1, IDR2, and IDR3). Bottom: Predictions of intrinsic disorder tendency of DDX10 using IUPred3 (https://iupred.elte.hu/), where scores above 0.5 indicate disorder. b Schematic representation of different truncated forms of mouse DDX10. c Droplet formation experiments evaluating 0.5 µM mCherry-tagged DDX10FL and truncated forms of DDX10: DDX10ΔIDR1, DDX10ΔIDR2, or DDX10ΔIDR3. Scale bars, 5 μm. d Droplet areas observed in panel (c). Data are presented as mean values ± SD. Significance was tested using the two-sided Mann-Whitney test. Exact p-values are reported in the figure. n = 10 fields. e Condensed fraction of DDX10-mCherry in experiments from panel (c). Data are presented as mean values ± SD with the indicated significance from two-sided t-test. Exact p-values are reported in the figure. n = 10 fields. f Live-cell images of mCherry-tagged DDX10 variants and Hoechst staining in NIH3T3 cells. Experiments were repeated three times independently with similar results. Scale bars, 5 μm. g FRAP experiments conducted on NIH3T3 cell lines. Fluorescence recovery curve obtained by bleaching a predefined spot in the nucleolus. Data are presented as mean ± SEM of n = 30 cells. h Brightfield images of DDX10-AID (+ OsTir1) mESCs overexpressing DDX10FL or different truncations treated with IAA at different time points (0 h, 24 h, and 48 h). Experiments were repeated three times independently with similar results. Scale bar, 200 µm. i Northern blot analysis of pre-rRNA intermediates in DDX10-AID (+ OsTir1) mESCs overexpressing DDX10ΔIDR1, DDX10ΔIDR2, DDX10ΔIDR3, and DDX10ΔIDR3-NLS, respectively, at 0 h and 48 h after IAA treatment. Experiments were repeated two times independently with similar results. j Northern blot analysis of the distribution of U3 snoRNA in the preribosome fraction of DDX10-AID (+ OsTir1) mESCs overexpressing DDX10ΔIDR1, DDX10ΔIDR3, and DDX10ΔIDR3-NLS, respectively, at 0 h and 48 h after IAA treatment. Experiments were repeated two times independently with similar results. Source data are provided as a Source Data file.