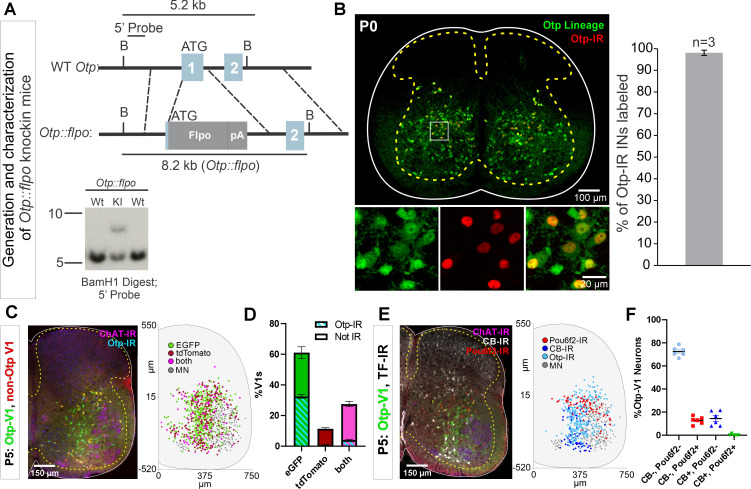
(
A) Targeting strategy to generate O
tp::flpo mice. Flpo was inserted into the ATG in the first exon of the Otp genomic locus. Dotted lines represent approximate regions of homology in the targeting vector. Southern blot (bottom) of BamH1-digested genomic DNA with a 5’ probe external to the targeting vector identifies a 5.2 kb wild-type fragment, and an 8.2 kb knock-in fragment. Not shown: Deletion of selectable neomycin resistance gene flanked by loxP sites by crossing to Protamine-Cre mice, which recombines the floxed PGK-Neo cassette in the male germline. (
B) Left, P0 lumbar spinal cord of O
tp::flpo, R26RCE.fsf-GFP mice, demonstrating expression in Otp-IR interneurons. Right, 98.1 ± 1.2% (mean ± SEM, n=3 mice) of Otp-expressing cells are labeled by the reporter. (
C) Intersection of
Otpflpo/+, En1cre/+ using the dual-color strategy with simultaneous expression of the Ai9 tdT and RCE-DC EGFP reporters. V1 cells that express
otp are labeled with EGFP (green). In these cells the Ai9-tdT reporter is effectively deleted by Flpo recombination dependent on the level of
Otp expression: strong (EGFP only) and weak (EGFP and tdT). V1 cells that express tdT only (red) never express
Otp. In addition, the sections were labeled with antibodies for Otp (light blue) to reveal cells that retained expression of Otp at P5 and choline acetyltransferase (ChAT) (deep blue) to localize the motor pools. Most V1s express EGFP, with tdT-only cells being a minority. Right, cell plot positions of some of the cell types identified in these sections (one mouse 6 ventral horns in L4/L5). Most cells are Otp-V1s and are shown here as green (EGFP only) and pink dots (EGFP and tdT). (
D) Quantification of cells with EGFP only (green), tdT only (red), or both (pink) (n=12 ventral horns from 2 mice; error bars show SD). 88.6% of V1s (1278 V1 cells analyzed in total) expressed EGFP; 60.8% were EGFP only, 27.8% were ‘yellow’ and surprisingly only 11.4% were tdT only. As expected, P5 Otp expression detected with antibodies was absent in most tdT-labeled V1 cells (93.5% of cells) and ‘yellow’ tdT+eGFP V1 cells (85.4%), but also in a significant proportion of EGFP-labeled V1 cells (46.7%). (
E) V1 cells transiently expressing Otp in embryo included cells of other clades. This was examined in
En1cre/+, Otpflpo/+,
R26RCE:dual-eGFP mice. Detection of Pou6f2, calbindin, and Otp in genetically labeled Otp-V1 cells at P5. Left confocal image and right cell plot (n=6 ventral horns from 1 mouse). Pou6f2-Otp-V1 cells concentrate in a dorsal band within the ventral horn. Calbindin-IR Otp1-V1 cells concentrate in the Renshaw region (ventral most region) and others are in the dorsal region of the ventral horn. Otp-V1 cells retaining Otp expression at P5 occupy all dorsoventral positions in the lateral spinal cord. (
F) Pou6f2 or calbindin immunoreactivity (-IR) in Otp-V1 cells (n=397) in 6 sections (each dot) in L4/L5 from one animal. Pou6f2 was detected in 12.8% of Otp-V1 cells at P5. In addition, 14.4% of Otp-V1 cells were calbindin+ and this included many in the Renshaw cell ventral region and few others located more dorsally. One rare dorsal Otp-V1 cell contained both Pou6f2 and calbindin (included in both percentages above). By limiting the analysis to ventral Otp-V1 interneurons in the Renshaw area we estimated that 8.3% of them are Renshaw cells. In conclusion, the Otp-V1 lineage includes cells from several V1 clades. Many downregulate Otp expression before birth, thus at P5 all Otp-expressing cells are restricted to the Foxp2-V1 lineage. In addition, many medial non-V1 Foxp2 cells also express Otp. Therefore, to specifically target the lateral group of proprioceptive Otp-Foxp2-V1 cells tightly associated to the LMC, a triple genetic intersection or alternatively, postnatally timed Otp-dependent recombination is necessary. Raw images of the blots and corresponding labeling are found in
Figure 7—figure supplement 1—source data 1 and
Figure 7—figure supplement 1—source data 2.