FIGURE 2.
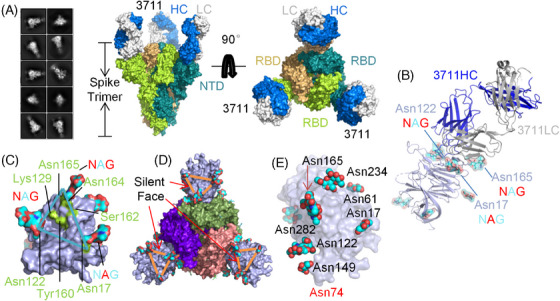
Overall conformation of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) spike N‐terminal domain “silent face” antibody 3711. (A) 2D classification, modeling, and Cryo‐EM map showing the surface of mAb 3711 in complex with SARS‐CoV‐2 spike extracellular domain (ECD) with two perpendicular views. The heavy chain and light chains of 3711 are colored blue and gray, respectively. The monomers of trimer spike protein (all receptor‐binding domains [RBDs] are in a closed state) are colored deep teal, limon, and wheat, respectively. (B) The overall conformation of mAb 3711 in complex with SARS‐CoV‐3 N‐terminal domain (NTD) in cartoon type. SARS‐CoV‐2 spike NTD is colored light blue in surface type; asparagine‐linked glycans are represented as cyan, blue, and red in spheres. (C) An epitope surrounded by Asn17‐, Asn122‐ and Asn165‐ linked glycans on NTD protein was designated as a “silent face”. Asparagine‐linked glycans are shown in cyan, blue, and red; the footprint of 3711 interacting as NTD is colored in limon. (D) The location of NTD “silent face” on trimer spike protein (PDB: 7C2L). Spike with three RBDs “down” is shown in the top view. NTD is colored light blue; three RBDs are shown in purple, smudge, and salmon, respectively. (E) Overall conformation as a surface type of SARS‐CoV‐2 NTD protein (PDB: 7C2L) with high glycosylation. Asparagine‐linked glycans are represented as cyan and red spheres. Asn74 linked glycans were undetected in 7C2L model. All protein structures were processed in PyMOL. NAG, N‐acetylglucosamine (GlcNAc).
