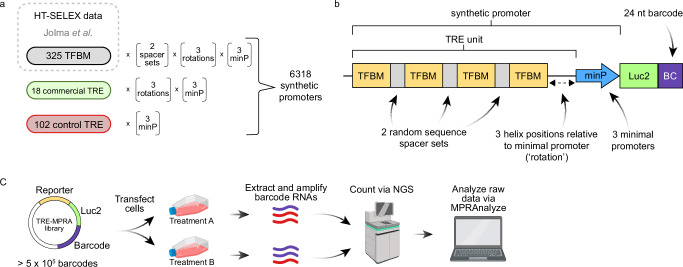
Fig. 1. TRE-MPRA overview.
a Transcription factor binding motifs (TFBMs) included in the TRE-MPRA library were derived from the HT-SELEX dataset of Jolma et al. Candidate synthetic promoters were designed by combining homotypic TFBMs and one of three minimal promoters in multiple configurations. Commercially available TREs, as well as negative control promoters were also included in the TRE-MPRA library. b Four copies of a given TFBM were oriented on alternating sides of the DNA double helix using spacer sets of random nucleotides and rotated along the helix relative to the minimal promoter (TRE unit). TRE units combined with minimal promoters (‘promoters’) regulate the expression of a Luc2 CDS containing 24 nucleotide barcodes in the 3’ UTR. Barcodes were mapped to associated TRE units using next-generation sequencing (NGS) during library preparation. c Barcoded RNAs were extracted from transfected cells and sequenced via NGS alongside the input TRE-MPRA plasmid libraries. Resulting barcode counts were used to generate estimated transcription rates and analyzed via MPRAnalyze to compare promoter activities between treatments. Created in BioRender. Zahm, A. (2024) https://BioRender.com/i28a348.