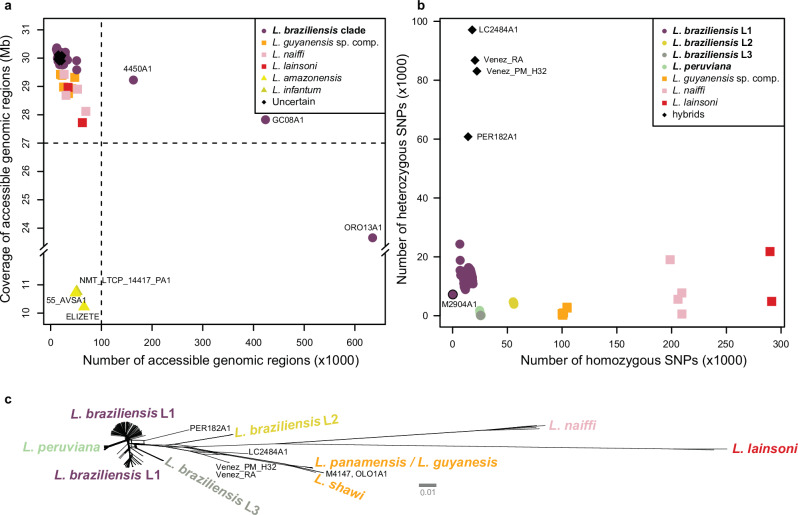
Fig. 1. Read coverage and natural genome variation in the Leishmania (Viannia) subgenus.
a Coverages across the accessible genomes of all 257 isolates. Isolates contained a median of 17.8 k accessible genomic regions, altogether spanning a median of 29.96 Mb (i.e. 91.5% of the haploid genome). Three isolates (55_AVSA1, ELIZETE, and NMT_LTCP_14417_PA1) were removed because of aberrantly low coverage of accessible regions (10.2-10.8 Mb) compared to the other isolates; in silico multi-locus sequencing analysis (MLSA) revealed that these isolates were Leishmania amazonensis (55_AVSA1, ELIZETE) and Leishmania infantum (NMT_LTCP_14417_PA1) (results not shown). Three other isolates identified as L. braziliensis (4450A1, GC08A1, and ORO13A1) were also removed because of low median coverages (9×–14×) and fragmented callable genomes. b Number of homozygous and heterozygous SNPs in the remaining 244 L. (Viannia) isolates. c Phylogenetic network of the 244 L. (Viannia) isolates based on 834,178 bi-allelic SNPs. Note: the bold legend labels in panels (a, b) represent the same isolates, all of the L. braziliensis clade.