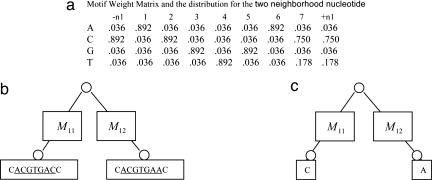
Fig. 3.
Update ancestor motif instance for gene met10 in yeast. (a) The weight matrix is constructed from all ancestral motif instances except the one in gene met10, which are sampled at the previous step. We also calculate the nucleotide distributions for the two neighboring nucleotides, which are the first and the last column in the weight matrix. (b) We have motif instances ACGTGAC for S. cerevisiae met10 and ACGTGAA for S. mikatae met10 (underlined) and their neighboring nucleotides from the previous sampling cycle. M11 and M12 are the substitution matrices at the corresponding branch. Here both of them are the same as defined in Table 1. (c) For each position, including the two neighboring positions, we calculate the probability that A, C, G, or T will appear there. Then one of the A, C, G, and T is drawn based on those probabilities for that position. For example, the probability that A will appear at the ninth position (the right neighboring position of the motif) is 0.036 × 0.8730 × 0.0182 and divided by some normalization constant, which results in 0.029.