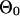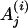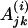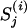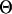|
Known parameters |
| m |
Number of current species; there are m – 1 ancestral species. |
| n |
Number of genes in one species. |
| I |
Species set. I = {0, 1,..., 2m – 2}. |
| S |
The set of regulatory sequences from all current species. |
|
Unknown parameters but can be estimated before applying the Gibbs Sampler |
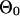 |
Background site model for species 0. |
| M0i |
Background substitution matrix at the ith branch of the tree, i = 0, 1,..., 2m – 3. |
| M1i |
Motif substitution matrix at the ith branch of the tree, i = 0, 1,..., 2m – 3. |
|
Unknown parameters that can be estimated from the Gibbs sampling |
| pi |
Probability of a gene containing motif instances in the ith species, i ∈ I.
|
| w |
Motif width. |
|
A(i)
|
Motif instance set in the ith species, i ∈ I.
|
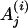 |
The motif instance in the jth gene of ith species, i ∈ I.
|
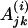 |
The kth nucleotide in the motif instance in the jth gene of the ith species, i ∈ I.
|
|
Notations that are used for describing the model and no particular interest |
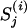 |
The regulatory sequence of the jth gene in the ith species, i ∈ I.
|
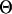 |
Motif model for species 0, the root species. |