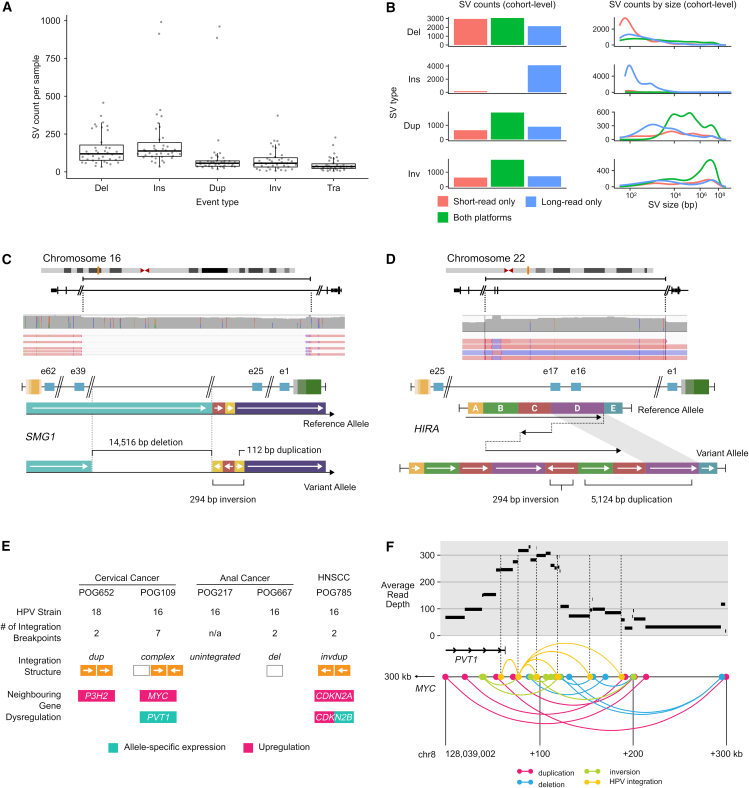
Figure 2.
Structural variants
(A) Per-sample counts of somatic SV calls in samples with matched normal (n = 43). Boxplots represent median, upper, and lower quartiles; whiskers represent 1.5× IQR. Del, deletion; Ins, insertion; Dup, duplication; Inv, inversion; Tra, translocation.
(B) Concordance of SV calling between platforms, summed across cohort.
(C) Schematic of a resolved complex foldback inversion affecting SMG1, including a deletion of exons 26–38, detected only in the nanopore data.
(D) Schematic of a resolved complex foldback inversion affecting HIRA, including duplication of exons 16–17, detected only in the nanopore data.
(E) Features of HPV integration characterized using nanopore sequencing in the five tumors with HPV.
(F) Diagram of a complex rearrangement (bottom) and alterations in read depth (top) involving HPV integration sites in a cervical cancer (POG109).