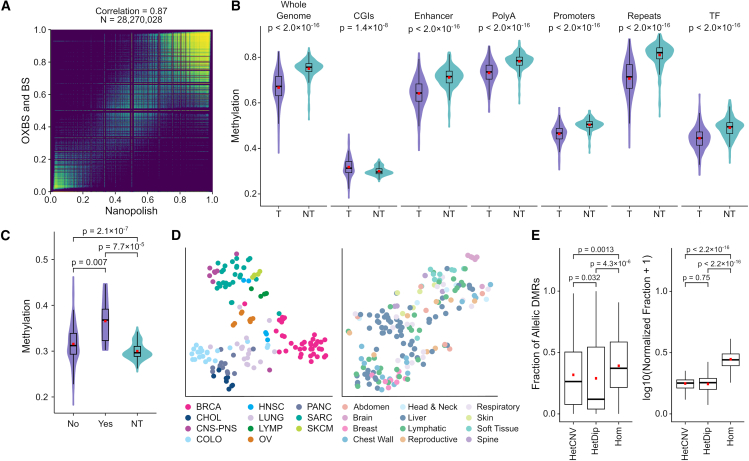
Figure 4.
Methylation
(A) Correlation of nanopolish methylation frequency with WGBS for POG044. BS, bisulfite sequencing; OXBS, oxidative bisulfite sequencing.
(B) Average methylation across tumors (T) compared with public WGBS methylation data from normal tissues and cells (NT), genome wide and at different genomic regions.
(C) Average methylation at CGIs in POG cases with either IDH-activating or TET-inactivating mutations (yes) compared with the remainder of the cohort (no) and public normal tissue (NT).
(D) tSNE plots based on DNA methylation at regulatory regions, compared with tumor type (left) and biopsy site (right).
(E) aDMR distributions by copy number (CN). Heterozygous diploid (HetDip) indicates CN-balanced regions. Heterozygous copy number variant (HetCNV) indicates CNV regions with both parental alleles. Homozygous (Hom) indicates LOH. All p values are Wilcoxon rank-sum test. Boxplots represent median, upper, and lower quartiles; whiskers represent 1.5× IQR.