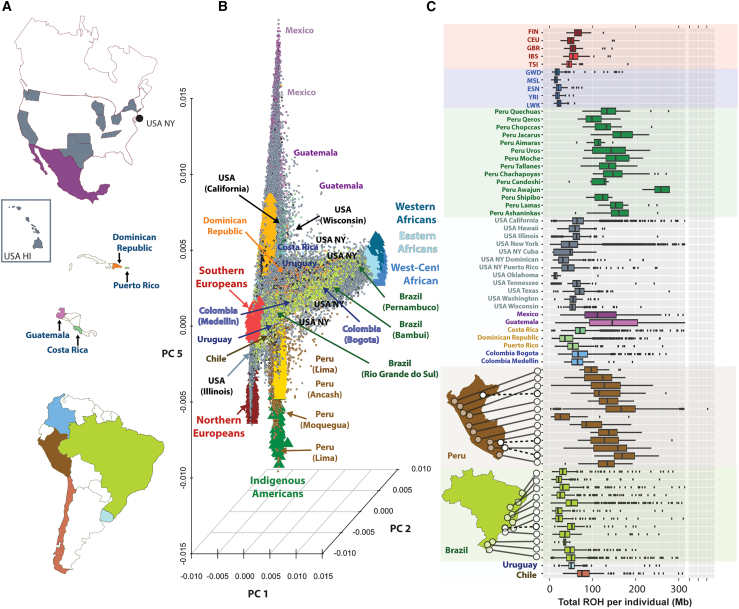
Figure 1.
Dimensionality reduction of genetic data and ROH for more than 53,000 unrelated LAms from the GLAD database
(A) Geographic distribution of GLADdb cohorts. Countries represented in GLADdb are highlighted with colors.
(B) PCA of the entire dataset based on high-quality imputed SNPs (Rsq >0.9) showing the sampling spread of LAms. Principal components 2 and 5 were plotted to show the axis of genetic diversity that explains the European-African (EUR-AFR) differentiation (PC2) and the diversity of Indigenous American ancestries from Mexico to Peru (PC5). All principal components are plotted in Figure S9.
(C) Distribution of genome-wide amount of ROHs for LAm groups and reference populations included in GLADdb. The upper part of the plot shows continental reference populations, and the lower part details the distribution in Peru and Brazil. Populations are sorted in a north-to-south pattern. This analysis was restricted to ROH segments >1 Mb. For patterns in ROH segments >8 Mb, see Figure S13. CEU, Utah residents with northern and western European ancestry from CEPH collection; ESN, Esan in Nigeria; EUR, European individuals; FIN, Finnish in Finland; GBR, British from England and Scotland; GWD, Gambian in Western Division-Mandinka; IBS, Iberian populations from Spain; LWK, Luhya in Webuye, Kenya; MSL, Mende in Sierra Leone; NAT, Indigenous American individuals; TSI, Toscani in Italia; USA HI, United States, Hawaii; USA NY, United States, New York; YRI, Yoruba in Ibadan, Nigeria.