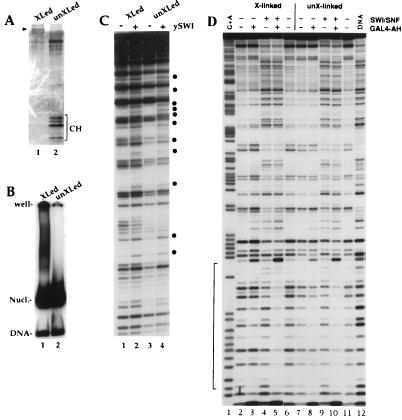
FIG. 6.
Histone octamer cross-linking does not affect nucleosomal DNA perturbation and enhanced GAL4-AH binding by SWI/SNF. Histone cross-linking conditions with dimethyl suberimidate and nucleosome reconstitution by transfer were described previously (31). (A) Sodium dodecyl sulfate-18% polyacrylamide gel showing that cross-linking conditions with dimethyl suberimidate create a protein band at around 100 kDa (arrowhead shows migration of 97.5-kDa marker) with no detectable free histone or partially cross-linked product (50 ng of histone loaded; 172-bp 5S DNA probe [21] used in the reconstitution). Note that the cross-linked histones take up less stain than control histones. The band that migrates more slowly than the core histones corresponds to bovine serum albumin, used to maintain stability. (B) Mobility shift gel showing that the cross-linked octamers do not significantly change the migration of the 5S mononucleosome. (C) Nucleosomes (25 nM) from the preparation used for panel B were incubated with 1 mM ATP in the presence (+) or absence (−) of purified SWI/SNF (2.5 nM) and analyzed by DNase I digestion. The bullets show where the intensity of a band is modified by SWI/SNF action. (D) A single-GAL4-site DNA probe (7) was reconstituted in nucleosome cores by using cross-linked (lanes 2 to 6) or non-cross-linked (lanes 7 to 11) histone octamers. The nucleosomes (25 nM) were incubated in the presence (+) or absence (−) of 15 nM SWI/SNF and/or 100 nM GAL4-AH dimers as indicated and were analyzed by DNase I digestion. Mg-ATP (1 mM) is also present in all of the lanes. The bracket indicates the GAL4-AH binding site. XLed and X-linked, cross-linked; unXLed and unX-linked, not cross-linked.