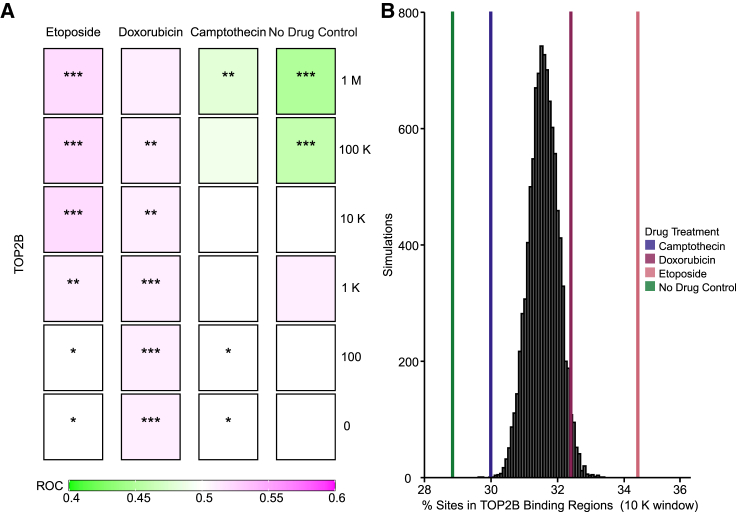
Figure 6.
AAV favors integration near inferred topoisomerase IIB binding sites selectively in the presence of the topoisomerase II poisons DOX and ETO
TOP2B binding sites were predicted by machine learning methods55 for HeLa cells. (A) Distribution of the AAV vector integration sites in the human genome relative to random controls (three times the number of sites as each experimental measure). TOP2B binding sites were predicted by machine learning. All biological replicates are collapsed by treatment condition, and associations were calculated using the ROC area method.35 Values of the ROC were scaled between 0.4 (negatively associated, green) and 0.6 (positively associated, purple). Significance was calculated by the ROC method (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). (B) Percentage of integration sites within 10,000 bp windows of inferred TOP2B binding sites. Random simulations are depicted in black and were generated from 1 million bootstraps of 1,000 randomly selected integration sites. Percent of sites in TOP2B tracks are presented as colored lines. No drug control (green), 62.5 nM of CPT (blue), 50 nM DOX (maroon), or 3.13 μM of ETO (pink).