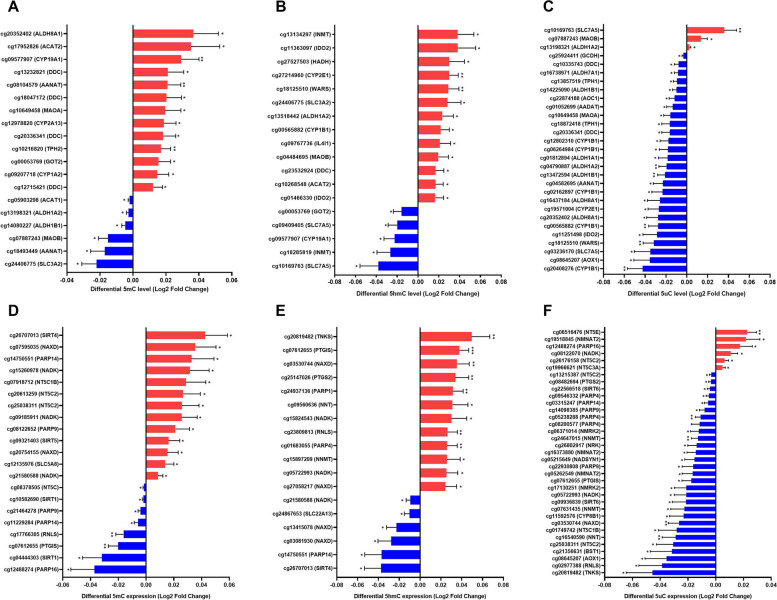
Fig. 2.
TRP and NAD pathway associated genes show significant differential DNA (hydroxy)methylation levels. Shown are significant CpG sites with its corresponding gene name in the tryptophan metabolic pathway (A-C) and NAD pathway (D-F) with up (red) or down (blue) regulation in the middle temporal gyrus of AD patients compared to age-matched controls. A, D Differential methylation (5mC) expression. (B, E) Differential hydroxymethylation (5hmC) expression. C, F Differential unmodified (5uC) expression. Differential expression is presented as log2 fold change. Differential regulation was assessed using limma differential DNA (hydroxy)methylation level analysis and p < 0.05 was considered significant. * p < 0.05 and ** p < 0.01. Error bars represent standard error of mean (SEM)