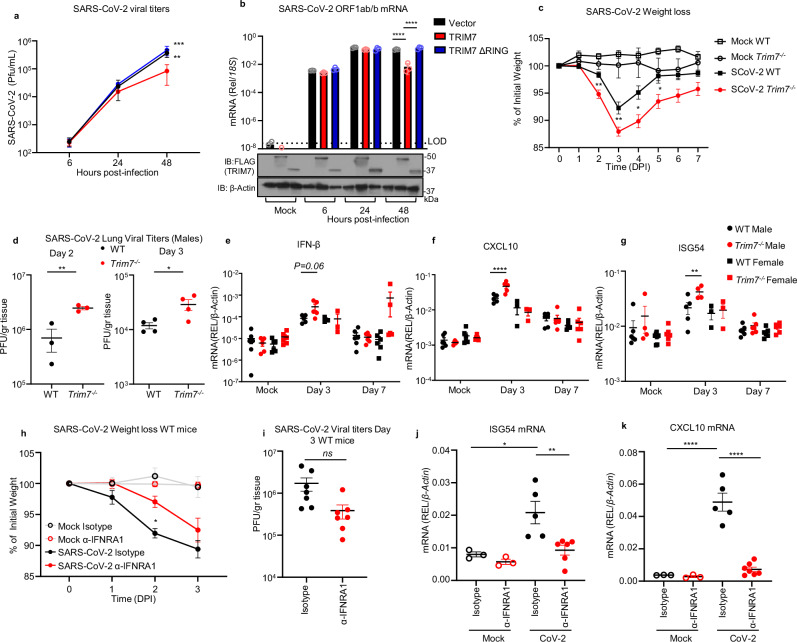
Fig. 2. TRIM7 has antiviral activity during SARS-CoV-2 infection.
a, b HEK293T-hACE-2 cells were transfected with 200 ng of TRIM7 WT or TRIM7-ΔRING followed by infected with SARS-CoV-2 (MOI 0.1). Viral titers quantified by plaque assay (a) or viral RNA by qPCR (b). Bottom (b) shows immunoblot control for TRIM7 expression. Two-way ANOVA Tukey’s multiple comparisons (**p = 0.0038, ***p = 0.0001, ****p < 0.0001). c C57BL/6NJ WT (n = 18, 9 females, 9 males) and Trim7−/− mice (n = 23, 10 females, 13 males) infected with maSARS-CoV-2 (1 × 106 PFU). Weight loss (c) data were from three combined independent experiments and multiple T-test (**p = 0.001, p = 0.003, *p = 0.005). d Viral titers (day 2, n = 3 males and day 3, n = 4 males per group). Two-tailed T-test (**p = 0.0083, *p = 0.03). e IFN-β gene expression in lung tissue by qPCR (n = 5 male day 3, 6 male day 7, and 3 female per infected group, mock n = 3 male and 3 female). f CXCL10 (same animal number as in (e). g ISG54 (mice numbers as in e, except for day 3, n = 5 WT and 4 Trim7−/− male). Two-way ANOVA Tukey’s multiple comparisons (****p < 0.0001, *p = 0.026). h–k IFNAR1 blockade: C57BL/6J WT (n = 7/group five females and two males) were treated with anti-IFNRA1 or isotype 24 h before infection with maSARS-CoV-2 (1×106 PFU). h Weight loss, two-way ANOVA Tukey’s multiple comparisons (*p = 0.023), i viral titers in the lung. j ISG54 gene expression by qPCR (n = 5 isotype, 6 α-IFNRA1 *p = 0.01, **0.009). k CXCL10 gene expression, one-way ANOVA Tukey’s multiple comparisons (n = 5 isotype, 7 α-IFNRA1, ****p < 0.0001). DPI days post infection, α-IFNRA1 anti-IFNRA1. Data were depicted as mean + SEM. Source data are provided in the Source data file.