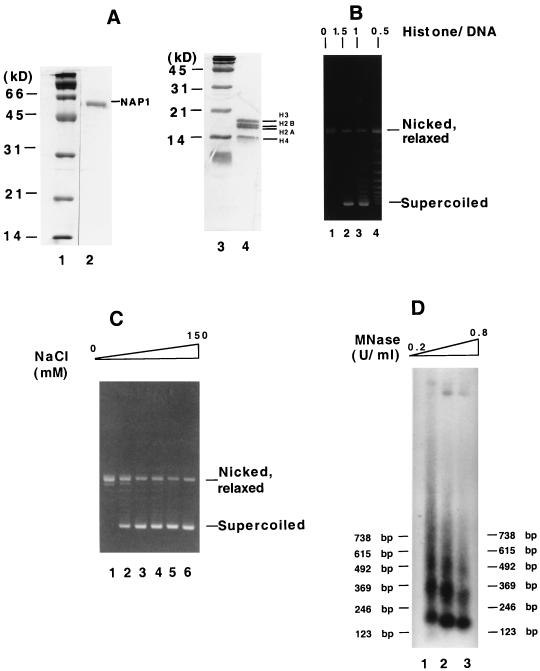
FIG. 1.
Reconstitution of chromatin in assays using purified proteins. (A) Analysis of purified proteins by SDS-PAGE (15% gel) and Coomassie blue staining. Lane 2, 3 μg of recombinant His6-tagged mouse NAP1; lane 4, 5 μg of human core histones; lanes 1 and 3, molecular weight markers (Bio-Rad). (B) DNA supercoiling assay for the optimum histone-to-DNA ratio. Chromatin was assembled with fixed amounts of histones and NAP1 but increasing concentrations (as indicated or the actual amounts) of relaxed circular DNA. (C) DNA supercoiling assay for the optimum salt concentration. Chromatin was assembled at NaCl concentrations of 0, 20, 25, 50, 100, and 150 mM. (D) MNase digestion analysis. Assembled chromatin was treated with increasing concentrations of MNase at room temperature. After deproteinization, the resulting DNA was resolved on a 1.5% agarose gel, transferred to nitrocellulose, and hybridized with a 5′-32P-labeled probe (5′TTCGGGGCGAAAACTCTCAAGGATCTTACCG3′) corresponding to plasmid sequence just outside the promoter region. The bands were visualized by autoradiography.