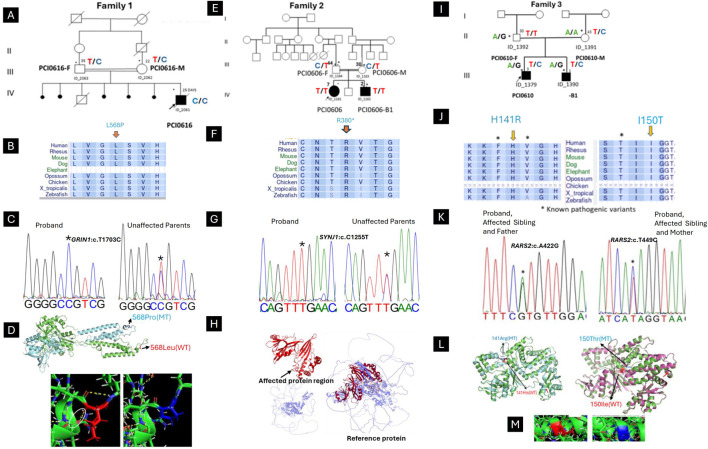
FIGURE 1.
Clinical and genetic findings of DEE patients (Family 1,2,3). (A) Pedigree of the case with neonatal myoclonic epilepsy showing recessive inheritance pattern (arrow indicates the proband and asterisks the participants seen in clinic). (B) The horizontal rectangle at the bottom represents the amino acid alignment around the mutated residue and their corresponding conservation among selected species using the UCSC Genome Browser. (C) Chromatogram showing the c.1703T > C variant in GRIN1. (D) Superimposed structure of GRIN1 wild type (green) and mutant (light blue), showing the shift of 3D conformational between. Hydrogen bond analysis showing the loss of bonding interaction of wild type leucine (white arrow) with phenylalanine in the mutant. (E) Pedigree of the case with infantile epileptic encephalopathy showing recessive inheritance pattern (arrow indicates the proband and asterisks the participants seen in clinic). (F) The horizontal rectangle at the bottom represents the amino acid alignment around the mutated residue and their corresponding conservation among selected species using the UCSC Genome Browser. (G) Chromatogram showing the (C) 1138C > T variant in SYNJ1. (H) Structure of the SYNJ1 reference protein, with the affected region highlighted in red. The truncated protein (in red) and the remaining portion of the reference protein (in light blue) illustrate the consequences of a premature stop codon, leading to the rupture of the full-length structure and the loss of both the α-helix and β-sheet regions. (I) Pedigree of the case with Ponto-cerebellar hypoplasia with seizures and showing recessive inheritance pattern (arrow indicates the proband and asterisks the participants seen in clinic). (J) The horizontal rectangle at the bottom represents the amino acid alignment around the mutated residue and their corresponding conservation among selected species using the UCSC Genome Browser.). (K) Chromatogram showing the compound heterozygous in RARS2 with both affected children (c.422A > G inherited from Father and c.449T > C inherited from Mother). (L) Superimposed structure of wild type RARS2 (green) and mutant (p.His141Arg; light blue) showing the partial loss of helix in the mutant (red arrow). (M) Superimposed structure of wild type RARS2 (magenta) and mutant (p.Ile150Thr; green) showing the partial loss of helix in the mutant (red arrow). Hydrogen bond analysis showing that there is not significant difference in bonding interaction between Isoleucine-150 and Threonine-150.