Abstract
The UK Biobank includes genotype information for about 500,000 patients for over 7000 phenotypes. However, owing to multiple testing correction for approximately 200 billion tests, many clinically and statistically significant associations remain unappreciated. We perform a focused analysis of the UK Biobank for 13 dermatologic conditions, including malignant melanoma, melanoma in situ, squamous cell carcinoma, basal cell carcinoma, actinic keratosis, seborrheic keratosis, psoriasis, lichen planus, systemic lupus erythematosus, hyperhidrosis, pilonidal cyst, sebaceous cyst, and lipoma. We identify 447 sentinel variants, which are enriched for protein-coding variants and an elevated combined annotation-dependent depletion (CADD) score compared with background variants. Through gene ontology enrichment analysis, we identify known pathways involved in melanoma, actinic keratoses, and squamous cell carcinoma and uncover additional pathways. We also uncover 5 protein-coding variants, which, to our knowledge, have not been previously reported, including LRP3 for lipomas, PLCD1 for sebaceous cysts, EIF3CL for lichen planus, TTK for pilonidal cysts, and MAPK15 for systemic lupus erythematosus.
Keywords: Dermatology, GWAS, Skin, UK Biobank
Introduction
GWASs link common variants to a phenotype of interest. Within dermatology, GWASs have provided important insights into psoriasis, skin pigmentation, and cutaneous malignancies, along with numerous additional traits (Fargnoli et al, 2006; Gudbjartsson et al, 2008; Stacey et al, 2009; Tsoi et al, 2012). More recently, meta-analysis GWASs have leveraged large cohorts of patients to identify loci associated with conditions, including melanoma (Landi et al, 2020).
The decreasing cost of sequencing has enabled large-scale efforts such as the UK Biobank to genotype and phenotype about 500,000 individuals for over 7000 phenotypes (Bycroft et al, 2018). However, owing to multiple testing corrections for approximately 200 billion tests, many clinically and statistically significant associations remain unappreciated.
We hypothesized that a focused analysis of the UK Biobank data would confirm well-established and identify new genetic associations that could provide insight into disease prediction and therapeutic targets. We filtered the UK Biobank GWAS data for dermatologic conditions (http://www.nealelab.is/uk-biobank/). Limiting the analysis to conditions with at least 500 individuals yielded 13 diseases: malignant melanoma, melanoma in situ, squamous cell carcinoma, basal cell carcinoma (BCC), actinic keratosis, seborrheic keratosis, psoriasis, lichen planus, systemic lupus erythematosus, hyperhidrosis, pilonidal cyst, sebaceous cyst, and lipoma.
Results and Discussion
Although GWASs are traditionally analyzed with a strict, fixed genome-wide significance threshold of 5 × 10−8, several groups have begun to reassess this cut off (Chen et al, 2021; Panagiotou et al, 2012; Schwartzman and Lin, 2011). We chose to pursue a less restrictive approach using the Benjamini–Hochberg procedure for several reasons. First, the set genome-wide significance cut off is not applicable because we are testing multiple traits. Second, we wanted to capture more putative variants than would be allowed with a strict Bonferroni correction, which would be set at 3.01e-10. Third, we consider a 5% false discovery rate (FDR), the expected proportion of type I error, in any potential association as acceptable. Fourth, although some true variants may be missed owing to the limitations of FDR under the linkage disequilibrium structure, this approach still allows for more putative variants than the aforementioned Bonferroni correction.
We filtered 166,373,136 associations with a Benjamini–Hochberg FDR of 5% to yield 447 sentinel variants (the most significant variant within a haplotype block) (https://ldlink.nci.nih.gov/?tab=snpclip). We defined a haplotype block as a 250-kb region with an R2 (a quantification of the strength of the nonrandom association between alleles at 2 different loci) >0.1. These are included in Supplementary Table S1. The sentinel variants had higher combined annotation-dependent depletion (CADD) scores, a predictor of deleteriousness, than background variants (4.55 vs 3.69, P = .0051) (Rentzsch et al, 2021). There was no statistically significant difference between PhyloP scores, a measure of conservation between 100 vertebrate species (0.03 vs 0.06, P = .6703). Although these variants may be deleterious, this analysis suggests that there has not been natural selection on the group of variants over the vertebrate lineage.
Next, we annotated the sentinel variants. They included 17 protein-coding variants (Table 1), 216 intergenic variants, 211 intronic variants, and 3 untranslated region variants. There is a higher frequency of protein-coding variants among sentinel than among background variants in the GWAS dataset (3.80% vs 0.90%, P < 1e-5). This analysis replicated variants previously identified in genes affecting skin pigmentation. Rs1126809 in TYR was associated with BCC and malignant melanoma (Gudbjartsson et al, 2008). Rs16891982 in melanoma antigen AIM1 (SLC45A2/AIM1) was associated with BCC (Stacey et al, 2009). Rs1805007 in MC1R was associated with actinic keratoses, melanoma in situ, and malignant melanoma (Fargnoli et al, 2006). Protein-coding associations, which we have not identified in previous literature, included variants in LRP3 with lipomas, PLCD1 with sebaceous cysts, EIF3CL with lichen planus, TTK with pilonidal cysts, and MAPK15 with systemic lupus erythematosus.
Table 1.
Protein Coding Sentinel Variants
| chr | Pos (hg19) | Raw P-Value | Trait | Gene | rsID | OR |
|---|---|---|---|---|---|---|
| chr16 | 89986117 | 5.62E-17 | Actinic keratosis | MC1R/TUBB3 | rs1805007 | 1.409451 |
| chr19 | 14006201 | 3.31E-06 | Actinic keratosis | BRME1 | rs148968932 | 7.381671 |
| chr5 | 33951693 | 4.93E-08 | Basal cell carcinoma | SLC45A2 | rs16891982 | 1.536028 |
| chr12 | 52913668 | 4.84E-13 | Basal cell carcinoma | K5 | rs11170164 | 1.374101 |
| chr11 | 89017961 | 3.09E-09 | Basal cell carcinoma | TYR | rs1126809 | 1.159397 |
| chr17 | 7571752 | 8.51E-07 | Basal cell carcinoma | TP53 | rs78378222 | 1.710695 |
| chr19 | 33696350 | 1.90E-07 | Lipoma | LRP3 | rs201590022 | 0.896641 |
| chr16 | 28403471 | 1.45E-06 | Lichen planus | EIF3CL | rs555978476 | 0.7403 |
| chr17 | 18158571 | 1.08E-06 | Melanoma in situ | FLII | rs138000313 | 21.41304 |
| chr16 | 89986117 | 1.34E-07 | Melanoma in situ | MC1R/TUBB3 | rs1805007 | 1.504559 |
| chr5 | 33951693 | 1.26E-08 | Malignant melanoma | SLC45A2 | rs16891982 | 1.744207 |
| chr11 | 89017961 | 1.29E-11 | Malignant melanoma | TYR | rs1126809 | 1.230106 |
| chr16 | 89986117 | 5.30E-27 | Malignant melanoma | MC1R/TUBB3 | rs1805007 | 1.683374 |
| chr6 | 80732138 | 1.55E-06 | Pilonidal cyst | TTK | rs56270911 | 26.49616 |
| chr3 | 98188784 | 1.89E-06 | Psoriasis | OR5K1 | rs201244755 | 217510 |
| chr3 | 38051211 | 3.93E-10 | Sebaceous cyst | PLCD1 | rs75495843 | 7.470784 |
| chr8 | 144804299 | 8.19E-07 | Systemic lupus erythematosus | MAPK15 | rs56038219 | 2.367418 |
Abbreviations: chr, chromosome; K5, keratin 5; Pos, Position.
We identified rs201590022, a protein-coding variant in LRP3, to be associated with lipomas. LRP3 is expressed in adipose tissue in both The Human Protein Atlas and GTEx (Genotype-Tissue Expression) datasets and is involved in the negative regulation of fat cell differentiation. However, we did not identify any known GWAS associations with this allele. We identified rs75495843, in PLCD1, to be associated with sebaceous cysts. PLCD1 was recently linked to trichilemmal cysts (Yousaf and Kolodney, 2022). We identified rs555978476, in EIF3CL, to be associated with lichen planus. Within the skin, EIF3CL is most highly expressed in granulocytes (The Human Protein Atlas, normalized transcript per million = 9.6). EIF3CL forms part of the elF3 complex that binds to the 40S ribosome and mRNAs to enable translation initiation. We identified rs56270911, in TTK, to be associated with pilonidal cysts. TTK is a critical mitotic checkpoint protein in which sequence variants have been associated with tumorigenesis (King et al, 2018). Finally, we identified rs56038219, in MAPK15, to be associated with systemic lupus erythematosus. MAPK15 enables MAPK activity and chromatin-binding activity and is involved in a variety of processes.
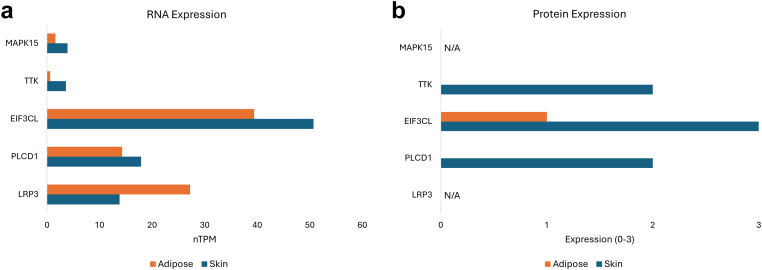
The 5 protein-coding genes mentioned earlier are expressed in the skin as either RNA or protein and are candidates for further validation (Figure 1). Of note, LRP1, which is involved in fat cell differentiation and associated with lipomas in this dataset, is more highly expressed in adipose tissue than in skin at the RNA level. Conversely, PLCD1, EIF3CL, TTK, and MAPK15 are all more highly expressed in skin than in adipose tissue at the level of RNA (Figure 1a). Although there are no protein expression data available for MAPK15 and LRP3, TTK, EIF3CL, and PLCD1 all have higher protein expression in skin than in adipose (Figure 1b).
Figure 1.
Expression of protein-coding genes in skin and adipose tissue from The Human Protein Atlas. (a) RNA expression in nTPM. (b) Protein expression data for each gene. No protein data were collected for MAPK15 or LRP3. The protein expression is annotated as not detected (0), low (1), medium (2), or high (3). This annotation is based on knowledge-based annotation. The annotation is based on the experienced evaluation of positive IHC signals in the 76 normal cell types analyzed. IHC, immunohistochemical; N/A, not applicable; nTPM, normalized transcript per million.
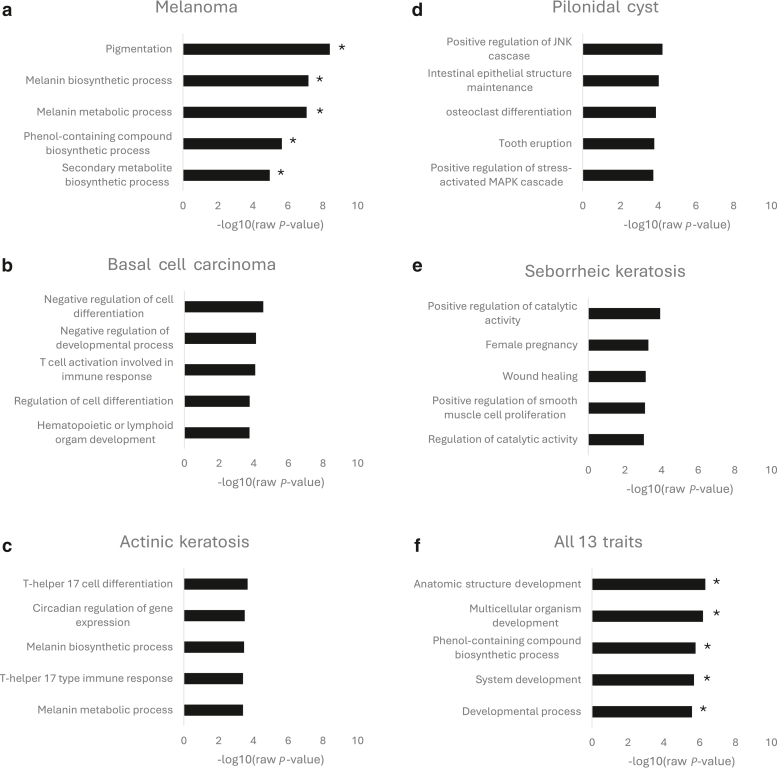
Next, we performed gene ontology enrichment for the nearest gene for each of the 447 sentinel variants (using the Gene Ontology Resource release 2022-03-22 [http://geneontology.org/]). Although we considered using more sophisticated tools for comprehensive enhancer-gene assignments, given the variety of phenotypes and cell lines of interest, we decided to perform the simplest and broadest approach, of the closest gene. Significant enrichments included pigmentation and melanin and phenol-containing biosynthetic processes with malignant melanoma (fold enrichments = 31.3, >100, and 48.03; FDR = 6.18e-5, 5.11e-4, and 6.79e-3) (Figure 2a). Other top enrichments that did not reach significance included T-cell activation for BCC (Figure 2b), T helper 17 response for actinic keratosis (Figure 2c), c-Jun N-terminal kinase cascade for pilonidal cysts (Figure 2d), and wound healing for seborrheic keratosis (Figure 2e). When all 13 traits were analyzed together, significant enrichments included anatomic structure development and phenol-containing compound biosynthetic process (fold enrichments = 1.46 and 11.4; FDR = 7.46e-3 and 8.65e-3) (Figure 2e).
Figure 2.
GO enrichment. (a–e) Top 5 enriched pathways for melanoma, basal cell carcinoma, actinic keratosis, pilonidal cyst, seborrheic keratosis. (f) Top 5 enrichments when all 13 traits were combined. Asterisk indicates FDR < 0.05. FDR, false discovery rate; GO, gene ontology.
Although T-cell activation for BCC did not reach significance, T-cell activation is a common mechanism of host defense against a variety of tumors. Some BCCs undergo immune-mediated regression, and infiltrating T cells have been identified in many BCCs. In addition, immune checkpoint inhibitors have shown effectiveness in early trials for BCC (Zilberg et al, 2023). If validated, this finding further supports the role for immune cell activation in management for BCCs. Similarly, the c-Jun N-terminal kinases, which was a top enrichment for pilonidal cysts in this analysis, contribute to inflammatory skin disorders, including dermal fibrosis, and have recently been implicated in the pathogenesis of renal cysts (Hammouda et al, 2020; Smith et al, 2021).
Although this analysis leverages a large biobank to identify putative variants associated with dermatologic conditions, it does have several limitations. First, to increase sensitivity, we use a Benjamini–Hochberg correction instead of the traditional Bonferroni correction used in GWAS studies. In addition, we do not include replication in an independent cohort but do find that many of the variants uncovered in this study have been previously reported in the literature. Nevertheless, further validation is required for these variants. Finally, we set a threshold of 500 cases for each trait analyzed in this manuscript. For many of these conditions, we will be underpowered to detect rare variants. This highlights the importance of continued expansion of large biobank datasets such as the UK Biobank and All Of Us Research Program.
In conclusion, we validated many known variants for dermatologic conditions and identified hundreds of variants, which we had not identified in previous literature, including 5 protein-coding sentinel variants. We highlight known pathways such as pigmentation and melanin for melanoma and T helper 17 for actinic keratoses and squamous cell carcinoma but also highlight several pathways such as c-Jun N-terminal kinase cascade for pilonidal cyst, wound healing for seborrheic keratosis, and proteoglycan synthesis for systemic lupus erythematosus, which may provide additional insight into these disease pathways. Finally, we provide a framework for performing focused studies with relevant clinical findings from within a larger biobank structure.
Materials and Methods
UK Biobank dataset
All analysis was completed on GWAS round 2 from the Neale lab (https://www.nealelab.is/uk-biobank/). These include imputed genotypes from HRC plus UK10 and 1000 Genomes reference panels as released by UK Biobank in March 2018. The imputed genotype data were performed by the UK Biobank Consortium, version 3 (https://biobank.ctsu.ox.ac.uk/ukb/label.cgi?id=100319). Quality control steps included in the UK Biobank analysis include that all variant sites with a call rate below 90% were filtered out, participants with sex chromosome aneuploidy were excluded, variants were also included if imputed INFO score (a metric that measures how well a variant has been imputed) >0.8, and variants were only retained if the allele count was at least 20. Additional quality control information can be found at https://pan.ukbb.broadinstitute.org/docs/qc/index.html#gwas-model. The dataset includes 7228 phenotypes and 16,131 genome-wide associations.
Statistical analysis
A total of 166,373,136 associations were filtered with a Benjamini–Hochberg FDR of 5% with Python Scipy to yield 447 sentinel variants. A sentinel variant was defined as the most significant variant within a haplotype block. A haplotype block was defined as 250 kb and an R2 > 0.1 using LDLink (https://ldlink.nci.nih.gov/?tab=snpclip). Combined annotation-dependent depletion scores and PhyloP scores were downloaded from the UCSC Genome Browser (hub_2140037_CADD_v1_6_hg19 and 100 vert cons (phyloP100wayAll). Background distributions were created using the shuf command to select a random 551 background variants from the initial dataset, and significance was tested with an unpaired t-test.
RNA and protein expression
RNA and protein expression data were obtained from the Human Protein Atlas. RNA data were pulled from the Consensus dataset (normalized expression transcript per million levels created by combining The Human Protein Atlas and GTEx transcriptomics datasets using the internal normalization pipeline). Protein data were pulled from the protein expression overview (knowledge-based annotation). For Figure 1, not expressed was quantified as 0, low was quantified as 1, medium was quantified as 2, and high was quantified as 3. This annotation has been performed by The Human Protein Atlas and is based on knowledge-based annotation. The annotation is based on the experienced evaluation of positive immunohistochemical signals in the 76 normal cell types analyzed. For a protein level to be included, it is necessary to have (i) an independent antibody targeting another epitope of the same protein, (ii) RNA-sequencing data, and (iii) available protein/gene characterization data. The profiles are performed using fixed guidelines on evaluation and presentation of the resulting expression profiles.
Gene ontology enrichment
Gene ontology enrichment was determined using the Gene Ontology Resource release 2022-03-22. The Gene Ontology Consortium’s gene ontology enrichment analysis tool does not correct for gene size bias. As described in the main text, the closest gene was determined for each SNP.
Data Availability Statement
Raw data for this analysis can be found at http://www.nealelab.is/uk-biobank/. All sentinel variants identified in this study are included in Supplementary Table S1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
ORCIDs
Jason C. Klein: http://orcid.org/0000-0001-9566-6347
Ruchika Mahapatra: http://orcid.org/0009-0005-7815-9841
Gary C. Hon: http://orcid.org/0000-0002-1615-0391
Richard C. Wang: http://orcid.org/0000-0003-4543-8295
Conflict of Interest
The authors state no conflict of interest.
Acknowledgments
This work was supported by National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin Diseases 5T32AR065969 to JCK and RCW. This work was presented at the 2022 Society for Investigative Dermatology Annual Meeting. The Sloan Kettering Open Access Publication Fund provided support for the article processing fee.
Author Contributions
Conceptualization: JCK, RCW; Functional Annotation: JCK, RM; Formal Analysis: JCK; Writing - Original Draft Preparation: JCK, GCH, RCW; Writing - Review and Editing: JCK, RM, GH, RCW
Declaration of Generative Artificial Intelligence (AI) or Large Language Models (LLMs)
The author(s) did not use AI/LLM in any part of the research process and/or manuscript preparation.
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2024.100322
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.xjidi.2024.100322.
Supplementary Material
Supplementary Table S1
References
- Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., et al. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Boehnke M., Wen X., Mukherjee B. Revisiting the genome-wide significance threshold for common variant GWAS. G3 (Bethesda) 2021;11 doi: 10.1093/g3journal/jkaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargnoli M.C., Altobelli E., Keller G., Chimenti S., Höfler H., Peris K. Contribution of melanocortin-1 receptor gene variants to sporadic cutaneous melanoma risk in a population in central Italy: a case-control study. Melanoma Res. 2006;16:175–182. doi: 10.1097/01.cmr.0000198454.11580.b5. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Sulem P., Stacey S.N., Goldstein A.M., Rafnar T., Sigurgeirsson B., et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma [published correction appears in Nat Genet 2008;40:1029] Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- Hammouda M.B., Ford A.E., Liu Y., Zhang J.Y. The JNK signaling pathway in inflammatory skin disorders and cancer. Cells. 2020;9:857. doi: 10.3390/cells9040857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.L., Zhang B., Li Y., Li K.P., Ni J.J., Saavedra H.I., et al. TTK promotes mesenchymal signaling via multiple mechanisms in triple negative breast cancer. Oncogenesis. 2018;7:69. doi: 10.1038/s41389-018-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi M.T., Bishop D.T., MacGregor S., Machiela M.J., Stratigos A.J., Ghiorzo P., et al. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat Genet. 2020;52:494–504. doi: 10.1038/s41588-020-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou O.A., Ioannidis J.P. Genome-Wide Significance Project. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41:273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman A., Lin X. The effect of correlation in false discovery rate estimation. Biometrika. 2011;98:199–214. doi: 10.1093/biomet/asq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.O., Jonassen J.A., Preval K.M., Davis R.J., Pazour G.J. C-Jun N-terminal kinase (JNK) signaling contributes to cystic burden in polycystic kidney disease. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey S.N., Sulem P., Masson G., Gudjonsson S.A., Thorleifsson G., Jakobsdottir M., et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi L.C., Spain S.L., Knight J., Ellinghaus E., Stuart P.E., Capon F., et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf A., Kolodney M.S. GWAS identifies three susceptibility loci for trichilemmal cysts. J Invest Dermatol. 2022;142:1221–1223.e5. doi: 10.1016/j.jid.2021.06.039. [DOI] [PubMed] [Google Scholar]
- Zilberg C., Lyons J.G., Gupta R., Damian D.L. The immune microenvironment in basal cell carcinoma. Ann Dermatol. 2023;35:243–255. doi: 10.5021/ad.22.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Data Availability Statement
Raw data for this analysis can be found at http://www.nealelab.is/uk-biobank/. All sentinel variants identified in this study are included in Supplementary Table S1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.