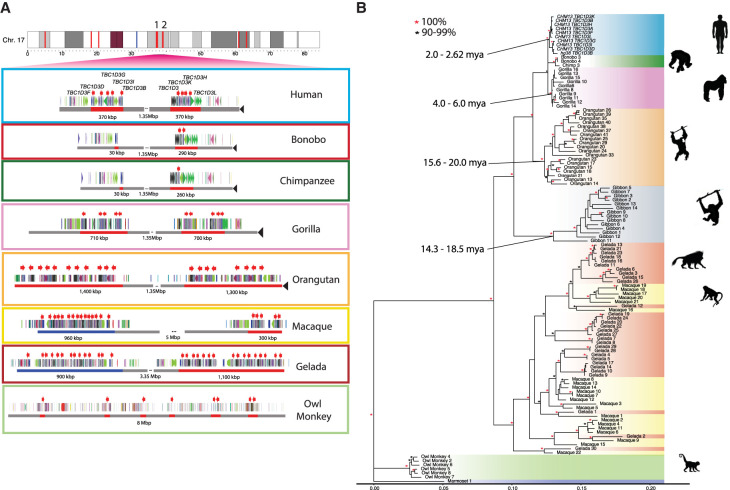
Figure 2.
Comparative genome structure and phylogeny of TBC1D3 gene family among primates. (A) TBC1D3 clusters 1 and 2 structure. Orthologous TBC1D3 clusters 1 and 2 are illustrated as two clustered regions (red blocks), with flanking unique sequence in gray for the primate lineages. Old World monkey TBC1D3 expansion 1, which is nonsyntenic, is highlighted in blue. TBC1D3 paralogs (red arrows) are embedded within other segmental duplication blocks, with DupMasker annotations illustrated with colored arrows. The diverse organizational differences of each expansion, including expansion size, duplicon content, and copy number, suggest independent expansion. (B) TBC1D3 neutral phylogeny generated by maximum likelihood; 2300 bp of intronic sequence were aligned between all primate TBC1D3 paralogs observed in A, with the marmoset sequence used as an outgroup. The phylogeny supports the hypothesis of independent expansion with the exception of the Old World monkeys (geladas and macaques) in which several copies duplicated before and after speciation of these two lineages (11 mya) (Liedigk et al. 2014).