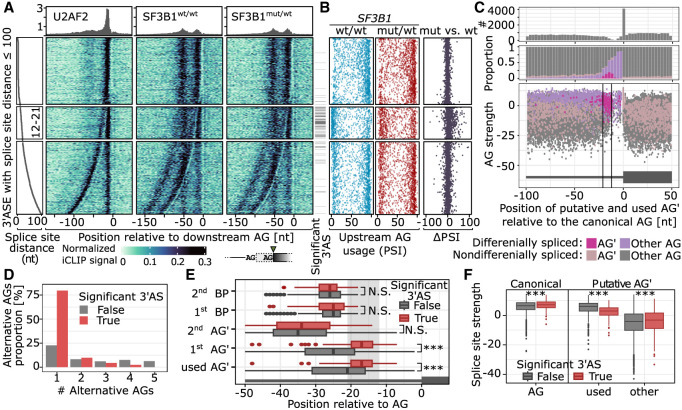
Figure 7.
SF3B1 promotes alternative proximal AG′ usage. (A) U2AF2 and SF3B1 binding to pre-mRNA based on the iCLIP signal. U2AF2 iCLIP was performed using K562-SF3B1wt/wt cells, and SF3B1 iCLIP was performed on K562-SF3B1wt/wt and K562-SF3B1K700E/wt. On the left panel, the splice-site distance is shown, followed by the iCLIP signal aligned to the more downstream AG used. The significance of the differential 3′AS usage between K562-SF3B1wt/wt and K562-SF3B1K700E/wt is shown as annotation bar on the right side of the iCLIP signal heatmap. (B) For every 3′AS from A, the upstream AG PSI value is denoted for SF3B1wt/wt (blue, left), SF3B1mut/wt (red, center), and the difference between SF3B1mut/wt and SF3B1wt/wt (black, right). (C) Distribution of AG occurrence (top), proportion of significantly alternatively used AG′ (middle), and AG′ scores among introns with significant (pink) or nonsignificant (violet) difference in usage between SF3B1mut/wt and SF3B1wt/wt (bottom). (D) Number of alternative AGs (AG′s) among regions with multiple AG′s. AG′s within 6 nt distance from AG were removed to avoid NAGNAG acceptor sites (Hiller et al. 2004). (E) Distance of AG′s and BPs from AGs among nonsignificant (gray) and significant (red) differentially splicing events. (F) Splice-site strength of canonical and alternative AGs as calculated with MaxEnt score (Yeo and Burge 2003). (A–F) Only 3′ASs with the following features were used: (1) placed chromosome scaffold, (2) classical AGs, (3) an intron became shorter in the mutant (the use of upstream alternative AG′), and (4) overlap with an iCLIP cross-link. The more used AG in SF3B1wt/wt was set as canonical AG.