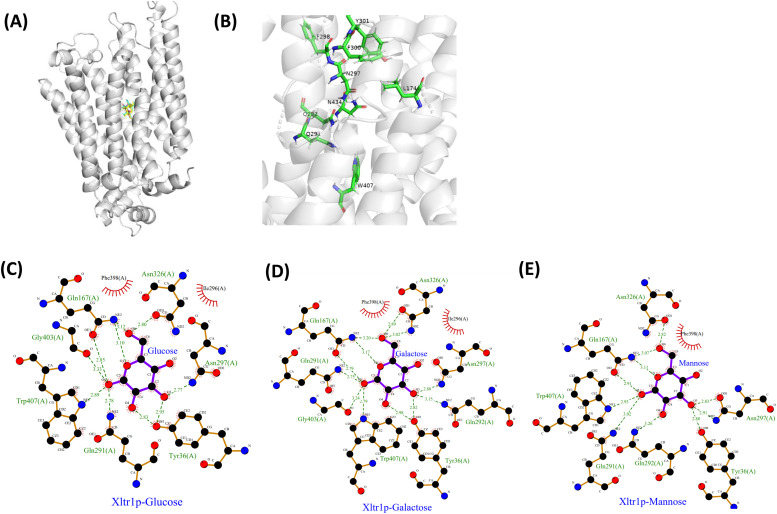
Fig. 2.
Substrate–protein docking analyses. (A) Homology modeling of Xltr1p based on the crystal structure of XylEp using SWISS-MODEL, followed by docking analysis with the substrates d-glucose (blue), d-mannose (magenta), and d-galactose (yellow) using Schrödinger software. (B) Display of the Xltr1p amino acid residues selected for mutation analysis. Analysis of the interactions between Xltr1p and d-glucose (C), d-galactose (D), or d-mannose (E) using Ligplot software.