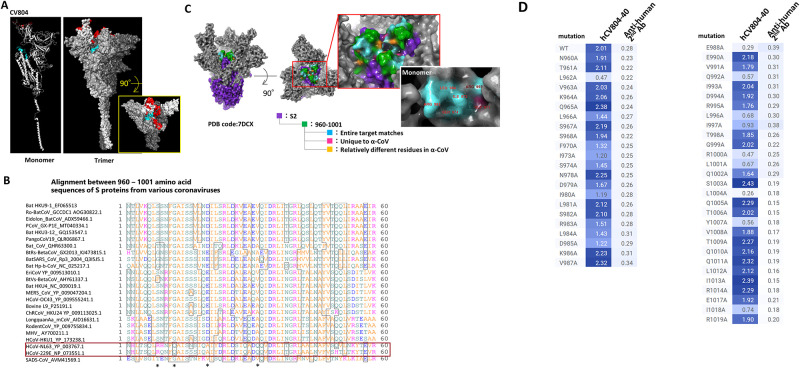
Fig 4. The epitope of CV804 is buried within the prefusion spike trimer.
We performed epitope mapping of the CV804 antibody binding to the spike protein of SARS-CoV-2 using HDX-MS. (A) The regions detected by HDX-MS were plotted onto the three-dimensional structure (PDB ID: 6vsb_1_1_1) shown in red for the ACE2 binding recognition domain and in cyan for the regions detected by HDX-MS. The figures from left to right depict the monomer and trimer, with the figure on the right showing a top view of the spike trimer. (B) Amino acid sequence alignment of the spike protein epitope candidate region (960–1001) was performed. The red box represents alpha coronaviruses, and the asterisks indicate amino acids that are relatively different in alpha coronaviruses. (C) Highly conserved regions in alpha and beta coronaviruses were extracted and mapped onto the 2-up structure of the Wuhan strain crystal structure (PDB code: 7DCX). Purple represents the spike 2 protein, green represents the detected epitopes, cyan represents the regions that are conserved among all target coronaviruses within the detected epitopes, pink represents residues that are unique to alpha coronaviruses, and yellow represents amino acid residues that are relatively different in alpha coronaviruses. The red box enlarges the central part of the trimer. The white box illustrates the monomeric state of the region indicated by the red box, with the amino acid residue numbers indicating the universally conserved regions among coronaviruses and exposed residues on the surface of the S2 protein. (D) Reactivity loss of CV804 with SARS-CoV-2 S protein in flow cytometry by substituting alanine for an amino acid in the S protein. 293T cells were transfected with full-length CoV-2 S protein expression plasmids with a point mutation designated, stained by humanized CV804-40, followed by PE-labeled secondary antibody. Binding activity is depicted in a heatmap of logMFI of PE signal.