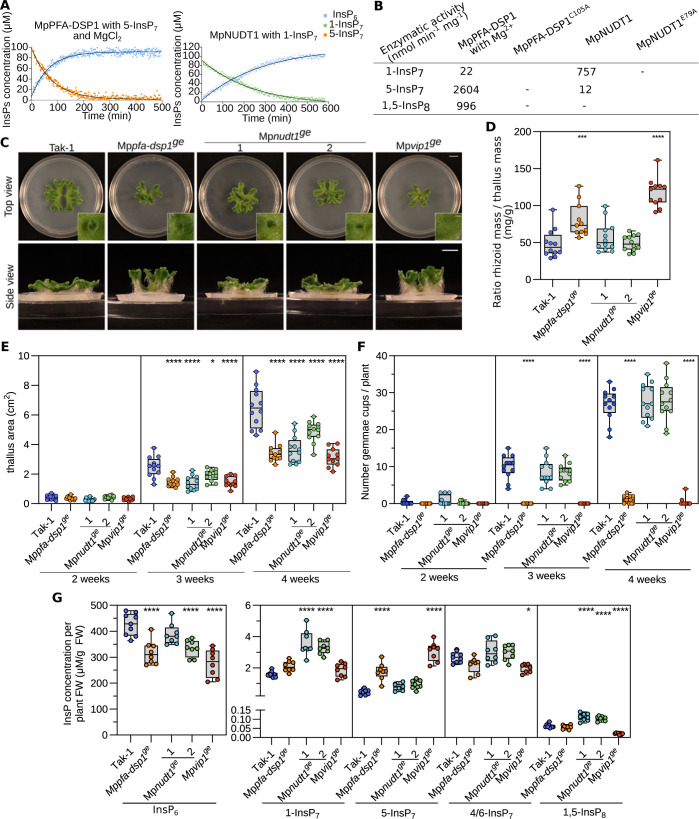
Fig 3. Inositol pyrophosphate phosphatases regulate Marchantia growth, development, and PP-InsP pools.
(A) Pseudo-2D spin-echo difference NMR time course experiments for MpPFA-DSP1 and MpNUDT1 inositol phosphatase activities, using 100 μM of [13C6]5-InsP7 or [13C6]1-InsP7 as substrate, respectively. (B) Table summaries of the enzymatic activities of MpPFA-DSP1 and MpNUDT1 vs. PP-InsPs substrates. (C) Representative top and side views of 4-week-old Tak-1, Mppfa-dsp1ge, Mpnudt1ge and Mpvip1ge plants. Plants were grown from gemmae on ½B5 plates in continuous light at 22°C. Scale bar = 1 cm. Single gemmae cups are shown alongside, scale bar = 0.1 cm. (D) Rhizoids mass normalized to thallus mass of 4-week-old Tak-1, Mppfa-dsp1ge, Mpnudt1ge and Mpvip1ge plants. Rhizoids were manually peeled with forceps. The weight of the rhizoids was normalized by the thallus weight of the same plant. Statistical significance was assessed with a Dunnett test with Tak-1 as reference (**** p < 0.001, *** p < 0.005, ** p < 0.01, * p < 0.05). (E) Thallus surface areas of Tak-1, Mppfa-dsp1ge, Mpnudt1ge and Mpvip1ge mutant lines in time course experiments. Plants were grown from gemmae on ½B5 plates in continuous light with 22°C and one plant per round Petri dish as shown in (C). For each genotype, 12 plants were analyzed. Statistical significance was assessed with a Dunnett test with Tak-1 as reference at each time point (**** p < 0.001, *** p < 0.005, ** p < 0.01, * p < 0.05). (F) Number of gemmae cups as a function of time for Tak-1, Mppfa-dsp1ge, Mpnudt1ge and Mpvip1ge. Statistical significance was assessed with a Dunnett test with Tak-1 as reference at each time point (**** p < 0.001, *** p < 0.005, ** p < 0.01, * p < 0.05). (G) InsP6 and PP-InsPs levels of 3-week-old Tak-1, Mppfa-dsp1ge, Mpnudt1ge and Mpvip1ge plants. (PP-)InsPs were extracted with titanium oxide beads and then quantified by CE-ESI-MS. Multiple comparisons of the genotypes vs. Tak1-1 were performed using a Dunnett test [105] as implemented in the R package multcomp [106] (**** p < 0.001, *** p < 0.005, ** p < 0.01, * p < 0.05).