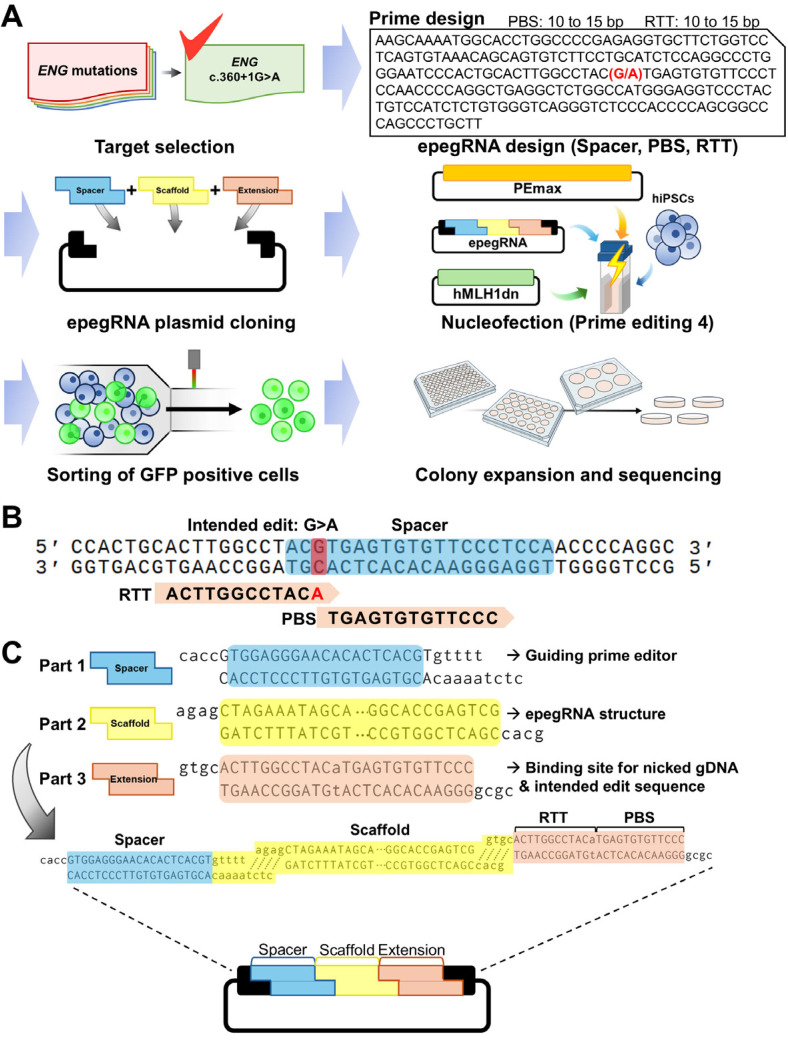
Fig. 2.
Strategy for prime editing to generate isogenic human induced pluripotent stem cells (hiPSCs) carrying clinically reported ENG mutation. (A) Scheme of the process for prime editing using hiPSCs. After choosing target mutation, engineered prime editing guide RNA (epegRNA) is designed using the PrimeDesign tool. The spacer sequence is preferably selected to be closest to the editing site, while reverse transcriptase template (RTT) and primer binding site (PBS) are typically chosen to be between 10 and 15 bp in length. To introduce designed sequences into epegRNA plasmid, annealed oligos for spacer, scaffold, extension including RTT and PBS are assembled into plasmid by golden gate cloning. Nucleofection is performed to introduce cloned epegRNA plasmid combined with PEmax plasmid and hMLH1dn plasmid, followed by isolating transfected cells via flow cytometry. Then, single-cell colonies are expended and sequenced to examine the genotype. (B) epegRNA design for targeting ENG c.360+1G>A mutation. The spacer sequence was selected as the closest to the target, and an A was added to the RTT sequence to change the target base to the desired sequence (G>A). (C) Scheme of Golden gate cloning for epegRNA plasmid construction. Oligo sets corresponding to spacer, scaffold, extension (RTT and PBS) sequence are shown.