Abstract
Mangroves are special vegetation that grows in the intertidal zone of the coast and has extremely high ecological and environmental value. Different mangrove species exhibit significant differences in ecological functions and environmental responses, so accurately identifying and distinguishing these species is crucial for ecological protection and monitoring. However, mangrove species recognition faces challenges, such as morphological similarity, environmental complexity, target size variability, and data scarcity. Traditional mangrove monitoring methods mainly rely on expensive and operationally complex multispectral or hyperspectral remote sensing sensors, which have high data processing and storage costs, hindering large-scale application and popularization. Although hyperspectral monitoring is still necessary in certain situations, the low identification accuracy in routine monitoring severely hinders ecological analysis. To address these issues, this paper proposes the UrmsNet segmentation network, aimed at improving identification accuracy in routine monitoring while reducing costs and complexity. It includes an improved lightweight convolution SCConv, an Adaptive Selective Attention Module (ASAM), and a Cross-Layer Feature Fusion Module (CLFFM). ASAM adaptively extracts and fuses features of different mangrove species, enhancing the network’s ability to characterize mangrove species with similar morphology and in complex environments. CLFFM combines shallow details and deep semantic information to ensure accurate segmentation of mangrove boundaries and small targets.Additionally, this paper constructs a high-quality RGB image dataset for mangrove species segmentation to address the data scarcity problem. Compared to traditional methods, our approach is more precise and efficient. While maintaining relatively low parameters and computational complexity (FLOPs), it achieves excellent performance with mIoU and mPA metrics of 92.21% and 95.98%, respectively. This performance is comparable to the latest methods using multispectral or hyperspectral data but significantly reduces cost and complexity. By combining periodic hyperspectral monitoring with UrmsNet-supported routine monitoring, a more comprehensive and efficient mangrove ecological monitoring can be achieved.These research findings provide a new technical approach for large-scale, low-cost monitoring of important ecosystems such as mangroves, with significant theoretical and practical value. Furthermore, UrmsNet also demonstrates excellent performance on LoveDA, Potsdam, and Vaihingen datasets, showing potential for wider application.
Keywords: Feature fusion, Mangrove species segmentation, Semantic segmentation, UAV remote sensing
Subject terms: Environmental sciences, Ocean sciences
Introduction
The mangrove ecosystem plays a key role in several aspects, such as coastline protection, biodiversity maintenance, and carbon storage1,2. Given the significant differences in ecological functions and environmental responses of different mangrove species, accurately identifying and segmenting various mangrove species are particularly important3. However, this task faces many challenges. First , mangrove species are highly similar in external morphology, such as branches, leaves, and crowns4. Second, mangrove forests usually grow in intertidal areas and marshes with complex lighting and backgrounds, and factors such as sunlight projection and water reflection can affect image quality5. Third, mangrove species demonstrate considerable morphological changes in different growth stages and generation periods6,7, and some small targets are easily lost in the segmentation process. Fourth, mangrove forests are susceptible to natural disasters, such as flooding and storms, resulting in tree damage and morphological changes8. All these factors together make the recognition of mangrove species based on images face great challenges. Mangrove forests grow in muddy areas that are difficult for humans to access9, and the lack of data and labeled samples increases the difficulty of developing intelligent recognition algorithms. With the continuous development of unmanned aerial vehicle (UAV) remote sensing technology and its related sensors, UAVs have shown great potential in improving the ability of mangrove species identification and monitoring10. They have the advantages of low cost, real-time data collection, and the provision of high-resolution images, which offer strong support for improving the management of coastal ecosystems11.
In recent years, benefiting from the continuous development of deep learning12,13, significant progress has been made in the field of semantic segmentation for UAV remote sensing14–16. Fully convolutional networks (FCNs)17 are remarkable works in introducing deep convolutional neural networks into the semantic segmentation task, their segmentation results are not fine enough and fail to fully consider the contextual information in images; meanwhile, the segmentation effect for mangrove tree species is poor, losing considerable fine information. U-Net18 is a U-shaped symmetric encoder-decoder network that fuses feature maps through skip connections to improve segmentation accuracy. While it preserves more details, it increases model complexity and computational cost. It may lose edge information during upsampling, affecting the precision of mangrove species segmentation, and struggles to cope with complex environments.In the field of traditional mangrove identification, Zhou et al.19 used a decision tree modeling algorithm to identify mangrove populations in the Zhangjiangkou Mangrove National Nature Reserve based on UAV hyperspectral images. Gao et al.20 utilized various integrated learning algorithms based on WorldView-2 hyperspectral satellite images to conduct a recognition study on the mangrove population in Gaoqiao Mangrove Reserve, Zhanjiang, Guangdong Province by using a dataset with a combination of various spectral features, such as texture features produced. However, these methods generally rely on hyperspectral images, their acquisition equipment is expensive and complicated to operate, and the cost of data processing and storage is high, which are not conducive to large-scale application and popularization. The success of DeepLab v3+21 stems from its Atrous Spatial Pyramid Pooling (ASPP) module22, which enhances multi-scale feature extraction capability. However, its feature fusion method is simplistic and fails to fully utilize information from different scales, resulting in insensitivity to mangrove target sizes and a large parameter count. Additionally, insufficient utilization of shallow features affects the segmentation of mangrove species boundaries and small targets, making it challenging to directly apply to mangrove species segmentation.UPerNet23 combines the Pyramid Pooling Module24 and Feature Pyramid Network25, improving segmentation accuracy through multi-scale feature fusion and adaptive aggregation. However, its complex structure and high computational overhead make it difficult to deploy in practice. In complex environments like mangrove forests, insufficient feature fusion leads to suboptimal recognition and segmentation results.Therefore, designing an efficient semantic segmentation network to realize accurate segmentation of mangrove species in high-precision UAV remote sensing mangrove images remains an enormous challenge with respect to the characteristics of mangrove species. Reducing the number of model parameters is also an important index to be considered to improve the efficiency of models and realize their actual deployment.
To solve the problems of the above methods, this study proposes a semantic segmentation network called UrmsNet for segmenting mangrove tree species in high-resolution UAV remote sensing RGB images, aiming to improve the accuracy and efficiency of ecological monitoring through efficient tree species segmentation techniques. UrmsNet first introduces spatial and channel reconstruction convolution (SCConv) and improves it to replace the standard convolution, which is more suitable for the task of semantic segmentation of mangrove tree species in UAV remote sensing images. It then introduces an adaptive selective attention module (ASAM) and a cross-level feature fusion module (CLFFM). ASAM extracts multi-scale features using multiple parallel adaptive selective feature modules (ASFM), which can capture the details of mangrove species in different sizes and morphologies to improve the discrimination of morphologically similar species. Afterward, the multiscale features are deeply fused via an adaptive context fusion module (ACFM), which adaptively selects useful context information and removes redundant information. This fusion approach enhances the network’s ability to characterize complex environments and reduces the effects of light and background complexity on recognition. CLFFM utilizes the detailed information in shallow features and the advanced semantic information in deep features. Shallow features are especially important for capturing small targets and boundary information, while deep features are helpful for overall semantic understanding. By comprehensively utilizing the above different layers of features, the module ensures that even small mangrove tree targets can be accurately segmented. On the mangrove tree species segmentation dataset based on UAV high-resolution RGB images established in this study, UrmsNet achieves superior segmentation results compared with other mainstream methods, with significant improvement in all evaluation indexes. The number of parameters and computational volume of the model are effectively controlled owing to the use of the improved SCConv+ to replace the standard convolution.
Nowadays, most mangrove tree community researchers use multispectral or hyperspectral images26–28, Yang et al.29 proposed a mangrove species classification model based on drone-acquired multispectral and hyperspectral imagery, using four machine learning models: AdaBoost, XGBoost, RF, and LightGBM for identification. The average accuracy reached 72% for multispectral imagery, while hyperspectral imagery achieved an average accuracy of 91%, with LightGBM method achieving the highest accuracy of 97.15%. Zhen et al.30 proposed a method based on the XGBoost algorithm for fusing image features, which comprehensively utilized three types of satellite remote sensing data (multispectral, hyperspectral, and dual-polarized radar satellite), ultimately achieving an identification accuracy of 94.02% for mangrove plant communities. The UrmsNet method proposed in this paper uses RGB images for mangrove species identification and segmentation, achieving mIoU and mPA of 92.21% and 95.98%, respectively, showing good results. Compared to methods using hyperspectral and multispectral data, it has the following advantages. UrmsNet utilizes ordinary RGB cameras for mangrove species identification, significantly simplifying the data collection process while substantially improving efficiency and reducing costs. Its compact data volume greatly alleviates storage and processing burdens, enabling more frequent ecological monitoring. UrmsNet lowers the technical threshold, maintains high compatibility with existing systems, and demonstrates potential for near real-time analysis. This approach is not only suitable for long-term monitoring but also easily integrates with other remote sensing data, providing comprehensive ecosystem information. Although it may be slightly inferior to traditional methods in terms of spectral information richness, UrmsNet exhibits excellent potential in mangrove population identification through advanced image processing and feature extraction techniques. These comprehensive advantages make UrmsNet a highly promising ecological monitoring tool, offering an efficient, economical, and practical solution for large-scale, long-term ecological studies.
In sum, this study provides a new, efficient, and accurate solution for the ecological monitoring and protection of mangrove forests, and the constructed dataset provides valuable data resources for research in related fields. The research results can provide technical support for the protection of mangrove forests and other important ecosystems and have important theoretical significance and application value. The main contributions are as follows.
Designing ASAM: ASAM extracts multiscale features of different mangrove tree species through parallel ASFM and deeply fuses these features through ACFM. ASFM improves the ability to discriminate morphologically similar tree species, while ACFM strengthens the network’s characterization ability and reduces the effects of light and background complexity on the recognition.
Designing CLFFM: CLFFM combines the detailed information in shallow features and the high-level semantic information in deep features to ensure that the boundary and small mangrove tree species targets can also be segmented accurately, which significantly improves the segmentation effect for mangrove tree species.
Constructing a high-quality UAV mangrove tree species segmentation dataset: In this study, a mangrove tree species segmentation dataset based on UAV high-resolution RGB images is constructed. The dataset covers several representative mangrove tree species in the Shankou Mangrove National Nature Reserve in Beihai City, Guangxi Zhuang Autonomous Region, China. This dataset provides not only a reliable experimental basis for this study but also valuable data resources for research in related fields and promotes the development of mangrove ecological monitoring and protection technology.
Methods
Overview of the overall architecture
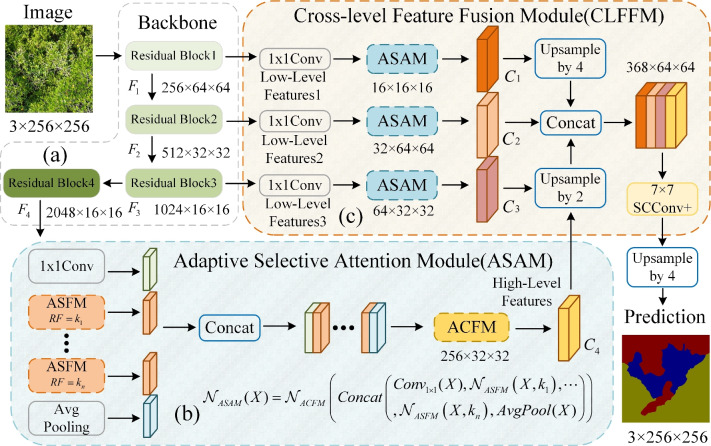
The overall structure of the proposed UrmsNet is shown in Fig. 1. The network adopts ResNet5031 as the backbone network for feature extraction, and the output feature maps of the backbone network are  ,
, 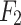 ,
, 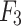 , and
, and 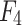 .ResNet50 is a classic and widely validated feature extraction network that performs excellently in tasks such as image classification and semantic segmentation. Its core advantage lies in effectively mitigating the vanishing gradient problem in deep networks through residual connections, thereby significantly improving the model’s training effectiveness and generalization ability. Additionally, ResNet50 has been pre-trained on large-scale datasets (such as ImageNet), providing a good starting point for our specific task. This transfer learning approach is particularly effective in tasks with limited labeled data, which aligns with our mangrove dataset situation. In comparison with other potential backbone networks, ResNet50 demonstrates unique advantages. Compared to VGG16/1932: Although VGG networks have a simple structure, they have a large number of parameters and high computational cost. ResNet50 has fewer parameters and lower computational requirements, improving computational efficiency while maintaining accuracy, making it more suitable for resource-constrained real-world deployment scenarios. Compared to Inception-v333: While Inception performs excellently in some tasks, its complex multi-branch structure may increase the difficulty of integration with our proposed SCConv+, ASAM, and CLFFM modules. ResNet50’s relatively simple structure is easier to integrate with custom modules. Compared to deeper ResNet variants (such as ResNet101): Although deeper networks may provide stronger feature extraction capabilities, they significantly increase computational overhead. Considering the characteristics of the mangrove segmentation task, ResNet50 offers a better balance between performance and efficiency. In summary, choosing ResNet50 as the backbone network for UrmsNet is based on a comprehensive consideration of accuracy, computational efficiency, and practical application requirements. This study improves SCConv34 with respect to the characteristics of the mangrove species segmentation task, redesigns the ASPP module on the basis of the improved SCConv and LSKNet35, and forms ASAM, which can enhance the network’s ability to characterize the multiscale targets of mangrove species at a small computational cost. This module can be used to cope with the morphological similarity and environmental complexity of mangrove forests, thus improving the recognition accuracy for mangrove tree species and effectively solving the problem of multiscale feature fusion in the ASPP structure, as well as the problem of high computational overhead caused by the increase in the memory consumption of large convolution kernels in LSKNet. CLFFM is designed to enhance the network’s utilization of information from the remaining features, especially boundary features and small-target features, to further improve the segmentation effect for mangrove tree species.
.ResNet50 is a classic and widely validated feature extraction network that performs excellently in tasks such as image classification and semantic segmentation. Its core advantage lies in effectively mitigating the vanishing gradient problem in deep networks through residual connections, thereby significantly improving the model’s training effectiveness and generalization ability. Additionally, ResNet50 has been pre-trained on large-scale datasets (such as ImageNet), providing a good starting point for our specific task. This transfer learning approach is particularly effective in tasks with limited labeled data, which aligns with our mangrove dataset situation. In comparison with other potential backbone networks, ResNet50 demonstrates unique advantages. Compared to VGG16/1932: Although VGG networks have a simple structure, they have a large number of parameters and high computational cost. ResNet50 has fewer parameters and lower computational requirements, improving computational efficiency while maintaining accuracy, making it more suitable for resource-constrained real-world deployment scenarios. Compared to Inception-v333: While Inception performs excellently in some tasks, its complex multi-branch structure may increase the difficulty of integration with our proposed SCConv+, ASAM, and CLFFM modules. ResNet50’s relatively simple structure is easier to integrate with custom modules. Compared to deeper ResNet variants (such as ResNet101): Although deeper networks may provide stronger feature extraction capabilities, they significantly increase computational overhead. Considering the characteristics of the mangrove segmentation task, ResNet50 offers a better balance between performance and efficiency. In summary, choosing ResNet50 as the backbone network for UrmsNet is based on a comprehensive consideration of accuracy, computational efficiency, and practical application requirements. This study improves SCConv34 with respect to the characteristics of the mangrove species segmentation task, redesigns the ASPP module on the basis of the improved SCConv and LSKNet35, and forms ASAM, which can enhance the network’s ability to characterize the multiscale targets of mangrove species at a small computational cost. This module can be used to cope with the morphological similarity and environmental complexity of mangrove forests, thus improving the recognition accuracy for mangrove tree species and effectively solving the problem of multiscale feature fusion in the ASPP structure, as well as the problem of high computational overhead caused by the increase in the memory consumption of large convolution kernels in LSKNet. CLFFM is designed to enhance the network’s utilization of information from the remaining features, especially boundary features and small-target features, to further improve the segmentation effect for mangrove tree species.
Figure 1.
Overall network architecture of UrmsNet.
In the process of image processing, the features of the input image are extracted by the backbone network, and four feature maps of different sizes are generated from 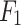 to
to 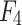 .
. 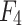 is processed by ASAM , and feature map
is processed by ASAM , and feature map 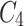 , which is rich in information on mangrove species, is extracted. Feature maps
, which is rich in information on mangrove species, is extracted. Feature maps 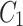 ,
, 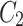 , and
, and 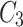 are obtained using CLFFM to process
are obtained using CLFFM to process 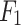 ,
, 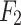 and
and 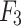 .Feature maps
.Feature maps 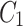 ,
, 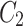 ,
, 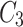 and
and 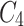 are fused and upsampled to generate the final accurate segmentation effect map.
are fused and upsampled to generate the final accurate segmentation effect map.
Convolution based on spatial and channel refinement (SCConv+)
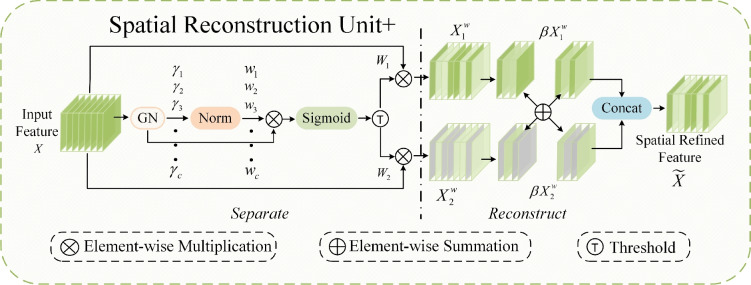
SCConv based on feature refinement is different from the standard convolution. It consists of two parts: the spatial reconstruction unit (SRU) and the channel reconstruction unit (CRU). SRU and CRU achieve feature refinement of redundant feature maps with low computational cost, thus enhancing the feature characterization capability.
SRU has two stages of separation and reconstruction, as shown in Fig. 2. The purpose of the separation stage is to separate the feature maps with rich spatial information from those with less spatial information. First, the scaling factor in the group normalization layer is used to evaluate the richness of spatial information of different feature maps. Then, it is mapped to the range of (0,1) by the sigmoid function to obtain the weights that measure the importance of spatial information at different locations in the feature map, which is gated by the threshold value (in this study, the threshold is set to 0.5). The whole process of obtaining weights in the separation stage is shown in (1).
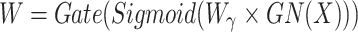 |
1 |
The purpose of the reconstruction stage is to generate combined features by adding features with large spatial information and features with small spatial information to save feature storage space. The cross-summing method is adopted to fully combine two weighted feature maps with different information. Feature maps 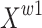 and
and 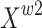 are summed up element by element in the channel dimension to obtain feature map
are summed up element by element in the channel dimension to obtain feature map 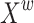 , which contains rich and refined spatial information. To further enhance the exchange of different information, this study increases the cross-summing ratio of information
, which contains rich and refined spatial information. To further enhance the exchange of different information, this study increases the cross-summing ratio of information 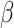 to control the exchange of information between feature maps
to control the exchange of information between feature maps 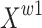 and
and 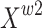 . This modulation can precisely control the information flow between different feature maps, thereby effectively capturing and distinguishing the subtle feature differences of mangrove species in different environments. The model can maintain high-precision segmentation effects in complex backgrounds and when faced with high similarity categories, improving the recognition accuracy for mangrove species. The whole process of reconstruction is shown below:
. This modulation can precisely control the information flow between different feature maps, thereby effectively capturing and distinguishing the subtle feature differences of mangrove species in different environments. The model can maintain high-precision segmentation effects in complex backgrounds and when faced with high similarity categories, improving the recognition accuracy for mangrove species. The whole process of reconstruction is shown below:
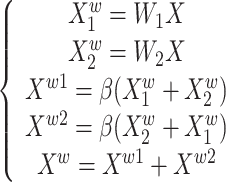 |
2 |
Figure 2.
Structure diagram of SRU+.
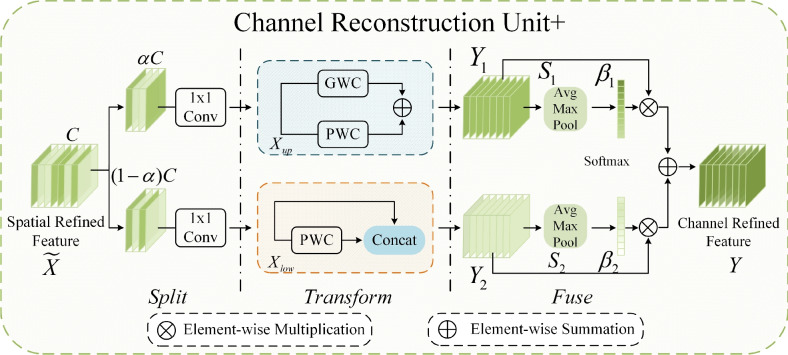
CRU has three stages, namely, split, transform, and fusion, as shown in Fig. 3. In the split stage, a spatially refined feature map is divided into two parts along the channel dimension, where 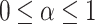 represents the proportion of segmentation. In the upper transform stage, group-wise convolution (GWC)36 and point-wise convolution (PWC)37 are used to replace the expensive standard convolution, extracting high-level semantic feature information and reducing computational costs, resulting in the output
represents the proportion of segmentation. In the upper transform stage, group-wise convolution (GWC)36 and point-wise convolution (PWC)37 are used to replace the expensive standard convolution, extracting high-level semantic feature information and reducing computational costs, resulting in the output  of the upper transform stage. In the lower transform stage, PWC and residual operations are used to extract hidden details from the feature map, serving as a complement to the rich feature extractor, leading to the output
of the upper transform stage. In the lower transform stage, PWC and residual operations are used to extract hidden details from the feature map, serving as a complement to the rich feature extractor, leading to the output  of the lower transform stage. In the fusion stage, feature maps
of the lower transform stage. In the fusion stage, feature maps 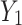 and
and 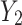 are adaptively fused, rather than simply added or concatenated. In this study, a combination of “Max Pool” and “Avg Pool” is used to collect comprehensive global spatial information of mangrove forests. By combining these two pooling techniques, the model can utilize the most salient features and global average information, which is especially effective when facing the complex mangrove forest environment. In mangrove forests, a large number of visual similarities exist among trees. Using a combination of these two different pooling strategies, the model can improve its ability to distinguish between different tree species and can effectively capture the global features and detailed information of mangrove tree species in different environments, thus enhancing the model’s ability to discriminate in complex backgrounds. Its computational process is shown as follows:
are adaptively fused, rather than simply added or concatenated. In this study, a combination of “Max Pool” and “Avg Pool” is used to collect comprehensive global spatial information of mangrove forests. By combining these two pooling techniques, the model can utilize the most salient features and global average information, which is especially effective when facing the complex mangrove forest environment. In mangrove forests, a large number of visual similarities exist among trees. Using a combination of these two different pooling strategies, the model can improve its ability to distinguish between different tree species and can effectively capture the global features and detailed information of mangrove tree species in different environments, thus enhancing the model’s ability to discriminate in complex backgrounds. Its computational process is shown as follows:
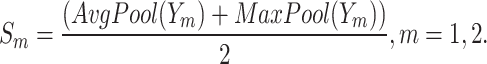 |
3 |
where 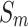 denotes the global spatial information of each channel of feature maps
denotes the global spatial information of each channel of feature maps 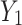 and
and 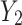 .
. 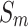 is processed using the softmax function to generate weight vector
is processed using the softmax function to generate weight vector 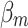 , which measures the importance of features and is calculated as
, which measures the importance of features and is calculated as
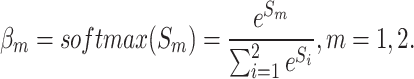 |
4 |
Finally, the feature importance weight vectors 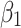 and
and 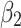 are weighted and added to the output feature maps
are weighted and added to the output feature maps 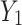 and
and 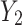 of the upper and lower transform stages, respectively, along the channel dimension, obtaining the final channel-refined feature map
of the upper and lower transform stages, respectively, along the channel dimension, obtaining the final channel-refined feature map 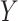 . The calculation process is shown in Eq. (5):
. The calculation process is shown in Eq. (5):
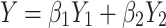 |
5 |
Figure 3.
Structure diagram of CRU+.
In this paper, the improved SRU and CRU are denoted as SRU+ and CRU+, respectively, and combined through spatial refinement, followed by channel refinement, to obtain the final SCConv+. Compared with the standard convolution, SCConv+ not only suppresses the redundant information of feature maps but also greatly reduces the computational overhead. Therefore, in this study, SCConv+ will be utilized to replace most of the standard convolutions in the subsequent designed modules to improve the model characterization capability while reducing the number of parameters and computation as much as possible.
Adaptive selective attention module (ASAM)
In remote sensing image analysis, especially in the task of semantic segmentation of mangrove species, distinguishing different tree species requires fine and complex feature extraction mechanisms. The visual differences between mangrove species may be subtle, and the distribution of tree species may be affected by complex and variable terrain and lighting conditions. To cope with these challenges, this study proposes ASAM, as shown in Fig. 1b. This module can enhance the network’s ability to focus on mangrove multiscale tree species targets with minimal computational cost to effectively address the morphological similarity of mangrove forests and the complexity of the environment, improve the recognition accuracy for mangrove tree species, and efficiently solve the problem of multiscale feature fusion in the structure of ASPP, as well as the problem of high computational overhead caused by the increase in the memory consumption of large convolution kernels in LSKNet.
ASAM is mainly divided into two major parts: multiscale feature extraction and feature fusion. In the multiscale feature extraction stage, 1x1 convolution, global average pooling, and multiple ASFMs are combined in parallel to capture the top-level features and their contextual information from different sensory fields. In addition, 1x1 convolution and global average pooling are used to supplement the global information. In the feature fusion part, the multiscale context information is spliced in the channel dimension and then deeply fused by ACFM to optimize the final feature representation. For input feature map 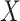 , the overall implementation flow of ASAM is
, the overall implementation flow of ASAM is
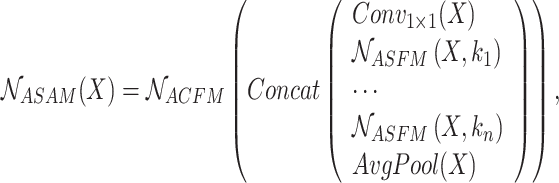 |
6 |
where input feature map 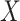 refers to output feature map
refers to output feature map 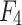 of the last stage of the feature extraction backbone network in the method in this chapter,
of the last stage of the feature extraction backbone network in the method in this chapter, 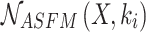 epresents the extraction of contextual feature information on different scales for input feature map
epresents the extraction of contextual feature information on different scales for input feature map 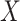 according to the setting of the sensing field size
according to the setting of the sensing field size 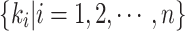 , and
, and 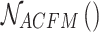 epresents the adaptive fusion of contextual feature information on different scales extracted by
epresents the adaptive fusion of contextual feature information on different scales extracted by 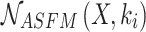 .
.
ASFM and ACFM are elucidated below.
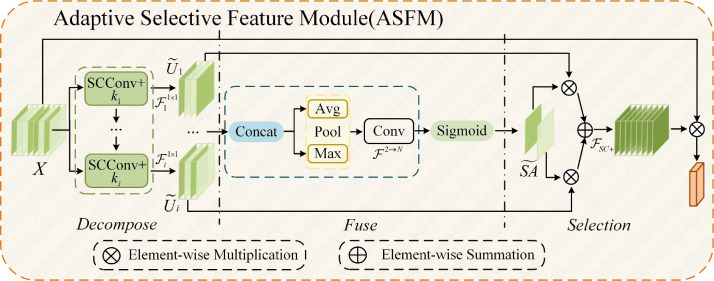
Figure 4 shows the ASFM structure, which contains three stages: decomposition, fusion, and selection. In the decomposition stage, SCConv+ is used instead of the standard convolution, and the large-sized convolution kernel is decomposed into multiple small-sized convolution kernels with different expansion rates. Thus, the overall receptive field remains unchanged, while the network has numerous choices of different sizes of receptive fields. A 1x1 convolutional layer 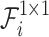 is obtained after each SCConv+, which is used to reduce the number of channels and thus the amount of the subsequent computation and the number of parameters and to obtain contextual feature
is obtained after each SCConv+, which is used to reduce the number of channels and thus the amount of the subsequent computation and the number of parameters and to obtain contextual feature 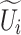 . This decomposition design can generate multiple features with different receptive fields, which makes the subsequent feature extraction flexible. In addition, the sequential decomposition has lower computational overhead than directly applying a single larger convolution kernel. While maintaining the same theoretical receptive field, the decomposition strategy greatly reduces the number of parameters compared with the standard large convolution kernel, which further lowers the computational cost of the network. In the fusion stage, context features
. This decomposition design can generate multiple features with different receptive fields, which makes the subsequent feature extraction flexible. In addition, the sequential decomposition has lower computational overhead than directly applying a single larger convolution kernel. While maintaining the same theoretical receptive field, the decomposition strategy greatly reduces the number of parameters compared with the standard large convolution kernel, which further lowers the computational cost of the network. In the fusion stage, context features 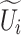 extracted from different sensory field convolution kernels in the decomposition stage are first subjected to a concatenation (Concat) operation in the channel dimension to aggregate the context information on different scales. Subsequently, channel-based average pooling and maximum pooling operations are performed on the spliced feature maps to obtain a representation of spatial relationships between different locations in the feature maps. The resulting feature maps of different spatial relations are spliced in the channel dimension to facilitate information interaction and fusion between different spatial relations, and the spliced feature maps (two-channel) are converted into two
extracted from different sensory field convolution kernels in the decomposition stage are first subjected to a concatenation (Concat) operation in the channel dimension to aggregate the context information on different scales. Subsequently, channel-based average pooling and maximum pooling operations are performed on the spliced feature maps to obtain a representation of spatial relationships between different locations in the feature maps. The resulting feature maps of different spatial relations are spliced in the channel dimension to facilitate information interaction and fusion between different spatial relations, and the spliced feature maps (two-channel) are converted into two 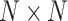 (N is the width of the feature maps) spatial attention maps using the convolutional layer
(N is the width of the feature maps) spatial attention maps using the convolutional layer 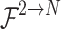 . These two attention maps reveal the interactions between different spatial relations and the weight distribution. Then, the sigmoid activation function is applied to obtain a spatial selection mask map corresponding to each convolution kernel in the decomposition stage. In the selection stage, the feature maps in the decomposed convolution kernel sequence are weighted in accordance with their corresponding spatial selection mask maps and then fused by the SCConv+ operation to obtain an attention feature map. The input feature maps are multiplied element by element with the attention feature maps to generate output feature maps. In this way, ASFM can adaptively adjust the size of the receptive field on the basis of different targets, which enables the network to extract the contextual information needed for different tree targets accurately, thus reducing the interference caused by insufficient contextual information and redundant background information.
. These two attention maps reveal the interactions between different spatial relations and the weight distribution. Then, the sigmoid activation function is applied to obtain a spatial selection mask map corresponding to each convolution kernel in the decomposition stage. In the selection stage, the feature maps in the decomposed convolution kernel sequence are weighted in accordance with their corresponding spatial selection mask maps and then fused by the SCConv+ operation to obtain an attention feature map. The input feature maps are multiplied element by element with the attention feature maps to generate output feature maps. In this way, ASFM can adaptively adjust the size of the receptive field on the basis of different targets, which enables the network to extract the contextual information needed for different tree targets accurately, thus reducing the interference caused by insufficient contextual information and redundant background information.
Figure 4.
Detailed structure diagram of ASFM.
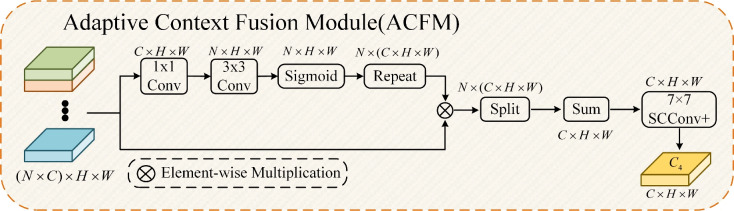
The detailed structure of ACFM is shown in Fig. 5. ACFM adopts a mechanism similar to attention to learn the importance of information at different levels but at the same spatial position, thereby retaining valuable feature information. First, ACFM uses two convolutional layers to reduce the channel dimension of the previously extracted feature map 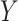 from
from 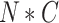 to
to 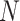 , where
, where 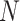 represents the total number of multiscale feature maps extracted by ASAM. Then, it uses the sigmoid activation function to obtain the importance weights representing the information at different positions of each layer’s feature map. Next, the obtained weights are replicated along the channel dimension to restore their dimensions to the original state and element-wise multiplied with
represents the total number of multiscale feature maps extracted by ASAM. Then, it uses the sigmoid activation function to obtain the importance weights representing the information at different positions of each layer’s feature map. Next, the obtained weights are replicated along the channel dimension to restore their dimensions to the original state and element-wise multiplied with 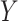 . After that, the result is sliced into
. After that, the result is sliced into 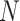 parts along the channel dimension and element-wise added at the channel level. Finally, feature refinement is performed through 7x7 SCConv+ to remove redundant information and retain effective information. Adaptive fusion is achieved to obtain the final output feature map
parts along the channel dimension and element-wise added at the channel level. Finally, feature refinement is performed through 7x7 SCConv+ to remove redundant information and retain effective information. Adaptive fusion is achieved to obtain the final output feature map 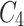 of ASAM. Through the above operations, ACFM can focus on the weights corresponding to the contextual feature information of each branch in ASAM in accordance with the importance of the contextual feature information of each branch. Adaptive fusion is realized on the basis of the weight ratio.
of ASAM. Through the above operations, ACFM can focus on the weights corresponding to the contextual feature information of each branch in ASAM in accordance with the importance of the contextual feature information of each branch. Adaptive fusion is realized on the basis of the weight ratio.
Figure 5.
Detailed structure diagram of ACFM.
Overall, ASAM combines 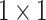 convolution, global average pooling, and multiple ASFM modules in a parallel manner to extract contextual feature information of mangrove species on different scales. Subsequently, the ACFM module is used to deeply fuse the extracted contextual feature information. ASAM enhances the network’s representation ability for different mangrove targets at a low computational cost. This approach can effectively address the morphological similarity of mangroves and the complexity of the environment, thereby improving the recognition accuracy for mangrove species. It also solves the problem of cumbersome multiscale feature fusion in the ASPP structure and the high computational overhead caused by the increased memory consumption of large convolution kernels in LSKNet.
convolution, global average pooling, and multiple ASFM modules in a parallel manner to extract contextual feature information of mangrove species on different scales. Subsequently, the ACFM module is used to deeply fuse the extracted contextual feature information. ASAM enhances the network’s representation ability for different mangrove targets at a low computational cost. This approach can effectively address the morphological similarity of mangroves and the complexity of the environment, thereby improving the recognition accuracy for mangrove species. It also solves the problem of cumbersome multiscale feature fusion in the ASPP structure and the high computational overhead caused by the increased memory consumption of large convolution kernels in LSKNet.
Cross-level feature fusion module (CLFFM)
To enhance the network’s comprehensive utilization of features from all layers, especially the information utilization of boundary features and small-target features, and to further improve the segmentation effect for mangrove tree species, this study designs CLFFM. With the increase in network depth, richer semantic features can be obtained, but this improvement is often accompanied with the loss of target details and boundary information. CLFFM ensures that small mangrove tree species targets and boundary information can be accurately segmented by utilizing the detailed information in shallow features and the advanced semantic information in deep features.
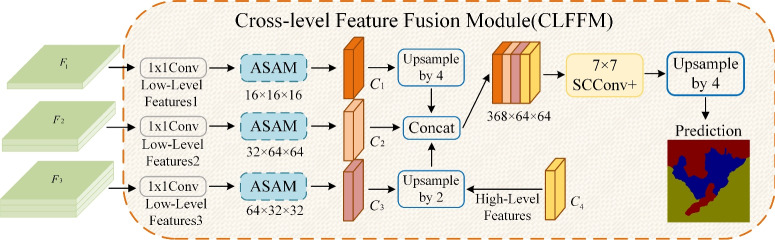
Figure 6 shows the structure of CLFFM, which contains three feature branches, each of which incorporates 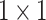 convolution and ASAMs. This design reduces the number of parameters and the computation amount and strengthens the ability of the shallow features to characterize the different sizes and shapes of mangrove trees. The shallow features cannot be compared with the deep features in terms of information abstraction and richness. In this study, the number of channels of the shallow features is reduced by
convolution and ASAMs. This design reduces the number of parameters and the computation amount and strengthens the ability of the shallow features to characterize the different sizes and shapes of mangrove trees. The shallow features cannot be compared with the deep features in terms of information abstraction and richness. In this study, the number of channels of the shallow features is reduced by 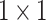 convolution, which not only decreases the computational overhead but also helps appropriately control the influence of the shallow features to ensure that they will not be adversely affected when fused with the deep features. This strategy is crucial for accurately depicting the complex boundaries and detailed features of mangrove forests, thus improving the accuracy and efficiency of the overall segmentation. For the three feature maps output from the backbone network, first,
convolution, which not only decreases the computational overhead but also helps appropriately control the influence of the shallow features to ensure that they will not be adversely affected when fused with the deep features. This strategy is crucial for accurately depicting the complex boundaries and detailed features of mangrove forests, thus improving the accuracy and efficiency of the overall segmentation. For the three feature maps output from the backbone network, first, 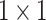 convolution is used to reduce the number of channels, and ASAM is adopted to enhance the characterization of the mangrove target. Then, upsampling and concatenation are performed with the feature maps obtained from the above step. Finally,
convolution is used to reduce the number of channels, and ASAM is adopted to enhance the characterization of the mangrove target. Then, upsampling and concatenation are performed with the feature maps obtained from the above step. Finally,  SCConv+ is used for feature refinement and upsampling to obtain the final semantic segmentation result.
SCConv+ is used for feature refinement and upsampling to obtain the final semantic segmentation result.
Figure 6.
Detailed structure diagram of CLFFM.
The clever structural design of CLFFM enables the network to effectively extract and utilize the key information in shallow, middle, and deep features, especially for the features of mangrove species targets. By combining with the aforementioned ASAM, CLFFM not only strengthens the ability to characterize the mangrove targets but also significantly improves the segmentation accuracy of the network for the different species of mangrove trees. This feature fusion strategy optimizes the overall effect of the features extracted from different depth levels and ensures that the model can capture the details of the mangrove targets and the complex background information to improve the accuracy and robustness of segmentation.
Experiment and analysis
Construction of datasets
RGB image acquisition based on UAV remote sensing images
In this study, we used a DJI Royal Series UAV equipped with a high-resolution RGB camera sensor as a monitoring tool to collect data on mangrove species in the Shankou Mangrove Forest National Ecological Nature Reserve in Beihai City, Guangxi Zhuang Autonomous Region, China. We acquired images at a flight altitude of 10 m and a forward speed of 3 m/s, with a total of 489 images with a resolution of 5280 3956 pixels. The RGB camera sensor was chosen because of its ability to provide high-resolution visual information on a UAV platform, along with the advantages of low cost and easy operation. Although RGB images are not as rich in spectral information as multispectral or hyperspectral images and lack information on additional bands, they have the advantages of easy acquisition and low cost of data processing. Through appropriate image processing and the feature extraction method presented in this paper, RGB images can be used to effectively identify mangrove populations. This approach not only reduces the cost of the study but also improves the maneuverability of data acquisition and is especially suitable for large-scale ecological monitoring and resource-limited areas.Although our data primarily comes from the Shankou National Mangrove Nature Reserve in Beihai City, Guangxi, this area exhibits typical characteristics of a subtropical mangrove ecosystem. It not only contains China’s main mangrove species but also faces environmental challenges and human disturbances similar to other mangrove regions. Additionally, our dataset encompasses plants at various growth stages, as well as samples affected by water submersion (mud-covered) and shaded specimens. Therefore, our dataset can reflect diversity to a certain extent. Furthermore, to further validate the model’s generalization ability and increase diversity, we have also introduced public datasets such as Potsdam, Vaihingen, and LoveDA for verification.
3956 pixels. The RGB camera sensor was chosen because of its ability to provide high-resolution visual information on a UAV platform, along with the advantages of low cost and easy operation. Although RGB images are not as rich in spectral information as multispectral or hyperspectral images and lack information on additional bands, they have the advantages of easy acquisition and low cost of data processing. Through appropriate image processing and the feature extraction method presented in this paper, RGB images can be used to effectively identify mangrove populations. This approach not only reduces the cost of the study but also improves the maneuverability of data acquisition and is especially suitable for large-scale ecological monitoring and resource-limited areas.Although our data primarily comes from the Shankou National Mangrove Nature Reserve in Beihai City, Guangxi, this area exhibits typical characteristics of a subtropical mangrove ecosystem. It not only contains China’s main mangrove species but also faces environmental challenges and human disturbances similar to other mangrove regions. Additionally, our dataset encompasses plants at various growth stages, as well as samples affected by water submersion (mud-covered) and shaded specimens. Therefore, our dataset can reflect diversity to a certain extent. Furthermore, to further validate the model’s generalization ability and increase diversity, we have also introduced public datasets such as Potsdam, Vaihingen, and LoveDA for verification.
Mangrove species selection and challenges
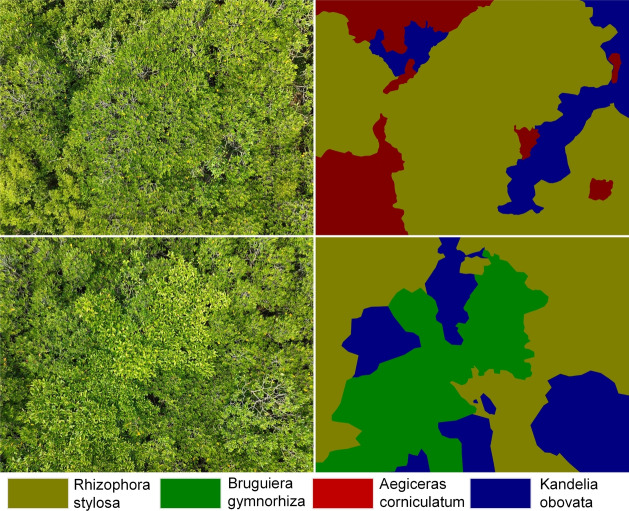
This research primarily focuses on four common mangrove species: Kandelia obovata, Aegiceras corniculatum, Bruguiera gymnorrhiza, and Rhizophora stylosa. As shown in Fig. 7, each species presents unique challenges in the segmentation process: Kandelia obovata: Leaves are elliptical with pointed tips and wedge-shaped bases. In densely grown areas, its canopy is easily confused with other species (like Aegiceras corniculatum). Seasonal changes (e.g., flowering period) affect its identification. Aegiceras corniculatum: Leaves are obovate or elliptical with rounded or slightly indented tips. Often intermixed with Kandelia obovata, making it difficult to distinguish in images. Its smaller size may be misidentified as background in high-resolution images. Bruguiera gymnorrhiza: Leaves are elliptical with acuminate tips and wedge-shaped bases. Its appearance varies greatly at different growth stages, increasing identification difficulty. Bruguiera gymnorrhiza in shaded areas is often misclassified. Rhizophora stylosa: Leaves are elliptical with acuminate tips and wedge-shaped bases, featuring distinctive prop roots. However, these root structures are not easily recognizable in top-view images, potentially leading to confusion with other species. Individuals growing near water edges are affected by tides, potentially causing significant morphological changes. Besides species-specific challenges, mangrove species segmentation faces some common difficulties: Morphological similarity: Different mangrove species are highly similar in appearance, increasing the difficulty of accurate classification. Environmental complexity: Mangroves grow in intertidal zones and swamps with complex lighting conditions and severe water reflection interference. Growth stage differences: The same species shows large morphological differences at different growth stages, increasing the difficulty of model generalization. Small target recognition: Seedlings and small-area distributed species are easily overlooked in the segmentation process. Natural disaster impacts: Typhoons, tsunamis, and other natural disasters may cause tree damage, altering their normal morphology. Data scarcity: Due to the unique growing environment of mangroves, obtaining high-quality, large-scale annotated data is challenging. Facing these challenges, drone remote sensing technology demonstrates unique advantages.
Figure 7.
Images of four mangrove species.
Preprocessing of acquired images
This paper used the LabelMe38 annotation tool under the guidance of mangrove species experts to conduct detailed annotations of different tree species (including Kandelia obovata, Aegiceras corniculatum, Bruguiera gymnorrhiza, and Rhizophora stylosa) in the images, as shown in Fig. 8. First, a team of experts consisting of two mangrove ecologists and one remote sensing specialist was assembled to develop annotation guidelines and oversee the entire annotation process. Detailed annotation guidelines were established, including identification features for each mangrove species, precise definitions of annotation boundaries, and unified rules for handling ambiguous areas and mixed species. Then, a two-tier annotation process was adopted: the first tier involved preliminary annotations by trained annotators, while the second tier involved review and correction by expert team members. Finally, 20% of the annotated images were randomly selected for cross-validation, with different experts re-annotating them to assess annotation consistency.To adapt to the input requirements of the deep learning model, in this study, the images of the selected region were cropped to 256  256 pixels, and a 32-pixel sliding-window method was used, from which 4692 images were cropped and selected. Of the images, 3285 were used for training, 471 for testing, and 939 for validation. Various data enhancement operations, including random image resizing, random cropping, random horizontal flipping, and photometric distortion, were used in this study to enhance the diversity of the data and the robustness of the model. This annotation and data augmentation process not only ensured the high quality and consistency of the dataset but also enhanced the effectiveness of model training. The dataset constructed through this method provides reliable training and testing resources for mangrove species segmentation research, while also laying a solid foundation for scientific research and application development in related fields.
256 pixels, and a 32-pixel sliding-window method was used, from which 4692 images were cropped and selected. Of the images, 3285 were used for training, 471 for testing, and 939 for validation. Various data enhancement operations, including random image resizing, random cropping, random horizontal flipping, and photometric distortion, were used in this study to enhance the diversity of the data and the robustness of the model. This annotation and data augmentation process not only ensured the high quality and consistency of the dataset but also enhanced the effectiveness of model training. The dataset constructed through this method provides reliable training and testing resources for mangrove species segmentation research, while also laying a solid foundation for scientific research and application development in related fields.
Figure 8.
Example images of mangroves with species labels.
Network training
The experimental environment for this study was a Ubuntu 18.04 operating system, and training and testing were conducted on a server equipped with an NVIDIA GeForce RTX4090 GPU (24 GB of video memory). The programming language used was Python 3.10, and the deep learning framework PyTorch 2.0.1+cu117 was employed.
In the parameter settings, we set the batch size to 2, which is a trade-off based on model complexity and GPU memory limitations. Although a smaller batch size may increase training time, it helps improve the model’s generalization ability. We use the Stochastic Gradient Descent (SGD) optimizer for training, with a momentum of 0.9 and weight decay of 0.0001. The momentum parameter helps accelerate convergence and reduce oscillations, while weight decay is used to prevent overfitting. The total number of iterations on the training set is 200 epochs, a value determined through preliminary experiments. We observed that validation set accuracy tends to stabilize after 180-190 rounds, allowing the model to fully learn the dataset’s features while avoiding overfitting. Additionally, we use a poly strategy to dynamically adjust the learning rate during training to achieve accelerated model convergence. The initial learning rate is set to 0.01 and gradually decreases as training progresses. This strategy has shown excellent performance in the semantic segmentation task of mangrove species, effectively balancing training speed and model performance. The learning rate changes over time according to the following formula:
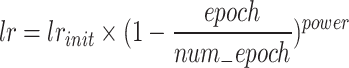 |
7 |
where lr is the new learning rate, 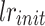 is the initial learning rate, epoch is the current training epoch number,
is the initial learning rate, epoch is the current training epoch number, 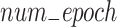 is the total number of training epochs. In our experiments, power is set to 0.9, which performed best in our tests. During the training process, we evaluate the model’s performance on the validation set every 5 epochs and save the best model. The entire training process takes approximately 20 h to complete on a single RTX4090 GPU. These parameters and training strategies ensure UrmsNet’s excellent performance in the mangrove species segmentation task while maintaining the model’s generalization ability and computational efficiency.
is the total number of training epochs. In our experiments, power is set to 0.9, which performed best in our tests. During the training process, we evaluate the model’s performance on the validation set every 5 epochs and save the best model. The entire training process takes approximately 20 h to complete on a single RTX4090 GPU. These parameters and training strategies ensure UrmsNet’s excellent performance in the mangrove species segmentation task while maintaining the model’s generalization ability and computational efficiency.
In large-scale ecological monitoring projects, efficient use of computational resources is crucial. Therefore, we conducted a detailed analysis of UrmsNet’s resource requirements during training and deployment stages:During the training phase, we used a server equipped with an NVIDIA GeForce RTX 4090 GPU (24GB VRAM) for training. On our mangrove dataset (approximately 4692 images with 256*256 resolution), a complete training (200 epochs) takes about 20 hours. For larger-scale datasets, we will adopt a distributed training strategy. Preliminary tests indicate that using 4 GPUs can reduce training time by about 60% without affecting model performance.In the deployment phase, on a single RTX 4090 GPU, UrmsNet can process about 80 images of 256*256 resolution per second. After using NVIDIA’s deep learning inference optimizer TensorRT, the inference speed can be increased by 2-5 times or more. At the same time, we consider that techniques such as quantization and knowledge distillation can be used to further compress the model size and improve model inference speed without significant loss of accuracy.
Evaluation metrics
To quantitatively assess the segmentation performance of the proposed network in this paper, we adopt Intersection over Union (IoU), mean Intersection over Union (mIoU), Pixel Accuracy (PA), mean Pixel Accuracy (mPA), number of Parameters, and Floating-Point Operations (FLOPs) as the measurement standards for the model.
Among these, the Intersection over Union (IoU) for each category and the mean Intersection over Union (mIoU) are the most classic evaluation metrics in semantic segmentation algorithms. Their calculation methods are shown in Equation 8,9:
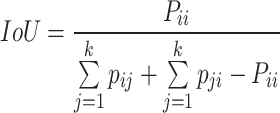 |
8 |
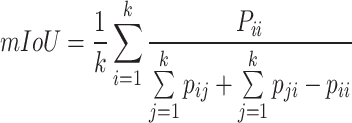 |
9 |
where k represents the total number of sample categories, 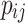 represents the number of pixels where the actual category of the sample is i while the network predicts category j ,
represents the number of pixels where the actual category of the sample is i while the network predicts category j , 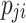 represents the number of pixels where the actual category of the sample is j while the network predicts category i , and
represents the number of pixels where the actual category of the sample is j while the network predicts category i , and 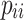 represents the number of pixels where both the actual category and the network-predicted category of the sample are i . By adopting IoU and mIoU as evaluation metrics for the model, we can accurately assess the overlapping regions of each category, thereby enhancing classification accuracy. This helps the model address challenges such as morphological similarities and environmental complexities when dealing with mangrove species, improving its identification capabilities.
represents the number of pixels where both the actual category and the network-predicted category of the sample are i . By adopting IoU and mIoU as evaluation metrics for the model, we can accurately assess the overlapping regions of each category, thereby enhancing classification accuracy. This helps the model address challenges such as morphological similarities and environmental complexities when dealing with mangrove species, improving its identification capabilities.
Pixel Accuracy (PA) is one of the indicators used to evaluate the overall accuracy of the model. It represents the proportion of correctly segmented pixels and is calculated by dividing the number of correctly segmented pixels by the total number of pixels. To comprehensively evaluate the segmentation results and accurately reflect the accuracy of all categories, the mean Pixel Accuracy (mPA) metric can be used. The calculation processes for PA and mPA are shown in Equation 10,11.
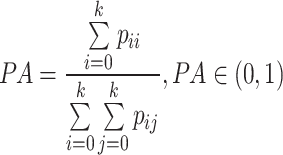 |
10 |
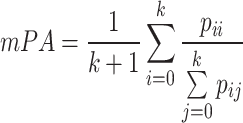 |
11 |
where 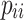 represents the number of correctly classified pixels, and
represents the number of correctly classified pixels, and 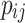 represents the number of pixels of class i predicted as class j . The range of PA is from 0 to 1, with values closer to 1 indicating better model performance. mPA calculates the average of all pixel accuracies, and compared to using PA alone, combining the evaluation results with mPA provides a more reliable assessment.d Using PA and mPA as measurement metrics for the model allows for an evaluation of the model’s overall recognition accuracy across all class pixels, thereby enhancing its ability to handle complex backgrounds and subtle boundaries when processing mangrove species.
represents the number of pixels of class i predicted as class j . The range of PA is from 0 to 1, with values closer to 1 indicating better model performance. mPA calculates the average of all pixel accuracies, and compared to using PA alone, combining the evaluation results with mPA provides a more reliable assessment.d Using PA and mPA as measurement metrics for the model allows for an evaluation of the model’s overall recognition accuracy across all class pixels, thereby enhancing its ability to handle complex backgrounds and subtle boundaries when processing mangrove species.
The number of Parameters and Floating-Point Operations (FLOPs) are the most commonly used evaluation metrics to measure the complexity of a network or algorithm. FLOPs represent the number of floating-point calculations required for one run of the network, while GFLOPs represent one billion floating-point operations. Generally, a higher value of FLOPs indicates that the network requires more computational resources and is more complex. The number of parameters represents the size of the network; typically, a larger number of parameters indicates a more complex network. Using Parameters and FLOPs as measurement metrics for the model allows for a comprehensive assessment of the model’s computational and storage requirements, thereby balancing accuracy with computational resources, optimizing performance, and promoting the practical deployment of mangrove species recognition technology.
Experimental results
Comparative experiments were conducted on the UAV mangrove tree segmentation number dataset established in this paper with the current representative methods to further evaluate the proposed network, and the experimental results were analyzed.
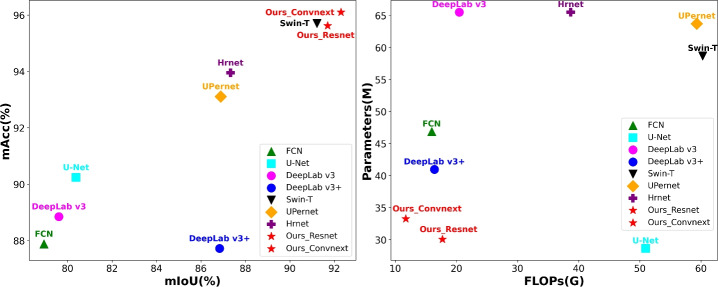
This study adopted IoU, Acc, Parameters, and Flops as evaluation metrics for the models. The experimental results of different models on the UAV mangrove species segmentation dataset are shown in Table 1 and Fig. 9. The proposed method achieved the best results in terms of mIoU for the categories of Aegiceras corniculatum, Bruguiera gymnorhiza, and Rhizophora stylosa. Although it performed slightly worse than Swin-Transformer39 in the Kandelia obovata category and mPA metric, the proposed model significantly reduced the parameters and flops compared with Swin-Transformer, giving it a greater advantage in practical deployment. Given that the backbone of the proposed network is ResNet50, when replaced with the more advanced ConvNeXt-Tiny40, it outperformed Swin-Transformer in all evaluation metrics, achieving the best results and further demonstrating the effectiveness of the proposed method.
Table 1.
Comparison of segmentation results of different models.
| Methods | IoU per class (%) | mIoU | mPA | Parame | Flops | |||
|---|---|---|---|---|---|---|---|---|
| K. obovata | A. corniculatum | B. gymnorhiza | R. stylosa | (%) | (%) | ters(M) | (G) | |
| FCN17 | 62.81 | 75.66 | 88.36 | 88.91 | 78.94 | 87.88 | 47.23 | 15.96 |
| U-Net18 | 72.65 | 75.55 | 84.74 | 88.58 | 80.38 | 90.24 | 28.99 | 50.94 |
| DeepLab v322 | 73.45 | 74.41 | 82.79 | 87.80 | 79.61 | 88.85 | 65.85 | 20.46 |
| DeepLab v3+21 | 79.77 | 85.73 | 89.86 | 91.99 | 86.84 | 87.72 | 41.32 | 16.40 |
| Swin-T39 | 87.30 | 91.05 | 92.69 | 93.84 | 91.22 | 95.70 | 59.02 | 60.28 |
| UperNet23 | 78.44 | 86.89 | 90.85 | 91.37 | 86.89 | 93.11 | 64.05 | 59.32 |
| HrNet41 | 81.91 | 84.73 | 90.82 | 91.88 | 87.33 | 93.95 | 65.85 | 38.69 |
| Ours-ResNet | 86.91 | 91.55 | 92.95 | 94.32 | 91.43 | 95.62 | 30.41 | 17.70 |
| Ours-ConvNext | 88.27 | 93.02 | 93.37 | 94.18 | 92.21 | 95.98 | 33.63 | 11.70 |
Figure 9.
Graphs of experimental results for different models.
The UrmsNet method proposed in this paper achieves the best results in terms of mIoU and mAcc, primarily due to the two modules we introduced: ASAM and CLFFM. Within ASAM, the ASFM achieves adaptive extraction and fusion of multi-scale features through three stages: decomposition, fusion, and selection. This provides a theoretical basis for addressing the morphological similarity of mangrove species, while offering new insights into feature selection theory in ecological image analysis.The ACFM within ASAM proposes a novel method for adaptive fusion of multi-scale features. By learning the importance of information from different layers but at the same spatial location, it achieves more effective feature fusion, providing new insights for solving multi-scale problems in remote sensing image analysis. ASAM effectively addresses issues such as morphological similarity and environmental complexity of mangrove species by combining the ASFM and ACFM modules. Meanwhile, CLFFM proposes a new cross-level feature fusion method by combining shallow and deep features. This method effectively utilizes detailed and boundary information from shallow features while retaining deep semantic information, providing new insights for solving scale issues and small target recognition problems common in mangrove and ecological monitoring.
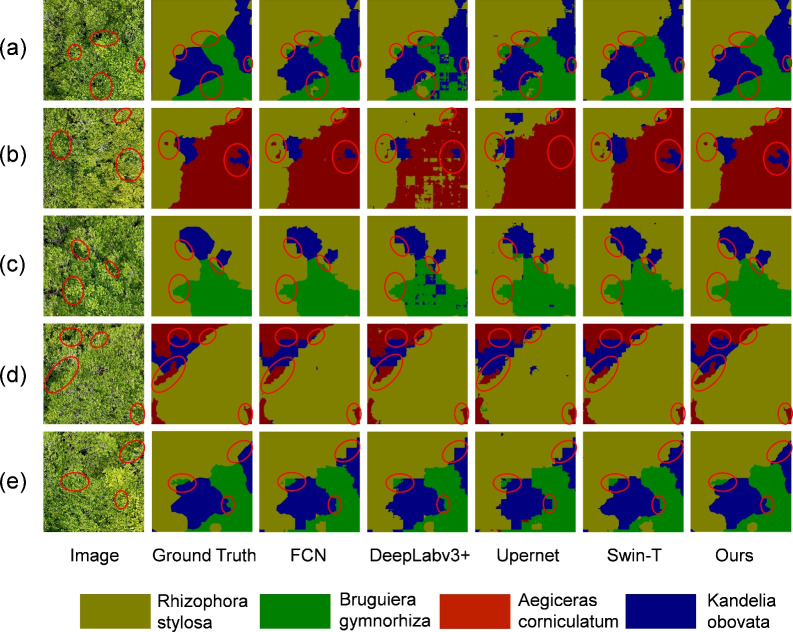
The results of the proposed method and several representative methods are shown in Fig. 10 to intuitively perceive the differences between them. From Fig. 10a, other methods could not accurately distinguish some tree species and had difficulty dealing with the morphological similarity of mangrove species. Figure 10b,d depict that other methods performed poorly when segmenting small-target mangroves. Figure 10c,e illustrate that other methods could not accurately distinguish the boundaries of mangrove species and even produce stair-like boundaries. By contrast , the proposed method performed better in solving the above problems. Compared with methods such as FCN, DeepLab v3+, Swin-Transformer, and UPerNet, the proposed method achieved more precise segmentation results and could better cope with the complexity and detail processing of mangrove species. Although UrmsNet performs excellently in most cases, it still faces challenges in certain specific situations. Through analysis of erroneous segmentation results, we identified the following scenarios where the model performs poorly or encounters difficulties: In areas where mangrove species are highly dense and multiple species grow in mixed patterns, the model sometimes struggles to accurately delineate boundaries between different adjacent species. This may be due to feature confusion caused by overlapping canopies. In areas with strong shadows or uneven illumination, the model’s segmentation accuracy decreases, indicating that the model’s adaptability to extreme lighting conditions still needs improvement. The model occasionally misjudges when identifying the same species at different growth stages, suggesting that the model needs to further enhance its ability to learn features across the growth cycle of tree species. These findings not only point out the current limitations of UrmsNet but also provide valuable directions for future improvements. By specifically enhancing data augmentation techniques, optimizing network structure, introducing multimodal data, or improving feature extraction methods, we hope to further improve the model’s performance in these challenging situations.
Figure 10.
Visualization comparison of segmentation results.
Ablation studies
In this section, the effectiveness of the proposed modules, including ASFM, ACFM, ASAM, and SDFFM, was evaluated. With DeepLabV3 as the baseline, the effectiveness of each module was verified by stacking them on top of the baseline. The results are shown in Table 2.
Table 2.
Ablation results of different modules.
| Methods | IoU per class (%) | mIoU | mPA | |||
|---|---|---|---|---|---|---|
| K. obovata | A. corniculatum | B. gymnorhiza | R. stylosa | (%) | (%) | |
| Baseline | 73.45 | 74.41 | 82.79 | 87.8 | 79.61 | 88.85 |
| Baseline+ASFM | 73.91 | 82.22 | 87.34 | 89.98 | 83.36 | 89.96 |
| Baseline+ACFM | 75.26 | 79.89 | 90.22 | 91.5 | 84.22 | 91.22 |
| Baseline+ASAM | 80.75 | 83.23 | 92.19 | 93.5 | 87.42 | 93.68 |
| Baseline+ASAM+SDFFM | 86.91 | 91.55 | 92.95 | 94.32 | 91.43 | 95.62 |
| Ours+Convnext-tiny | 88.27 | 93.02 | 93.37 | 94.18 | 92.21 | 95.98 |
From the results in Table 2, after ASFM was added, the mIoU increased by 3.75%, and the mPA increased by 1.11%. The IoU of each category, including K. obovata, A. corniculatum, B. gymnorhiza, and R. stylosa, also improved to a certain extent, indicating that ASFM can improve the segmentation performance of mangrove species to a certain degree. When ACFM was integrated, the mIoU increased by 4.61%, and the mPA increased by 2.37%, demonstrating the effectiveness of introducing ACFM. When ASFM and ACFM were combined into ASAM, the mIoU increased by 7.81%, and the mPA increased by 4.83%, proving the effectiveness of ASAM. Adding SDFFM on top of ASAM increased the mIoU by 11.82% and the mPA by 6.77%, achieving a significant improvement and the best results. The effectiveness of the proposed modules was verified through the above ablation experiments. When the ResNet50 backbone was replaced with ConvNeXt-Tiny, the accuracy still improved to a certain extent, further demonstrating the applicability of the proposed modules.
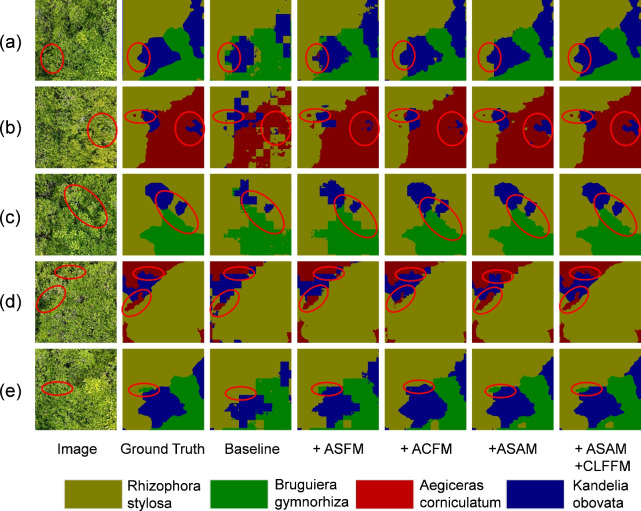
To intuitively perceive the effectiveness of the proposed modules, this study visualized the results of individual modules and the stacking of multiple modules. Through this approach, the contribution of each module to the improvement of model performance can be clearly demonstrated. As shown in Fig. 11, in the baseline method, the model exhibited certain limitations when segmenting mangrove species, struggling to accurately distinguish subtle differences between species and resulting in large areas of misclassification and significant blurring at the boundaries. When ASFM or ACFM was introduced, the model performed finely in handling species details, and the misclassified areas were significantly reduced; however misclassification still occurred at the boundaries. When ASFM and ACFM were combined into ASAM, the model’s performance was further improved, and it could accurately distinguish various mangrove species. However, it still fell short in handling small-target mangroves and species boundaries. When SDFFM was added on top of ASAM, the fusion of global and local features became effective, the segmentation of small-target mangroves was precise, the ability to identify subtle details was significantly enhanced, and the processing of species boundaries became clear, avoiding blurring and misclassification phenomena. This finding further verified the effectiveness of the proposed modules in this paper.
Figure 11.
Visualization comparison of ablation experiments.
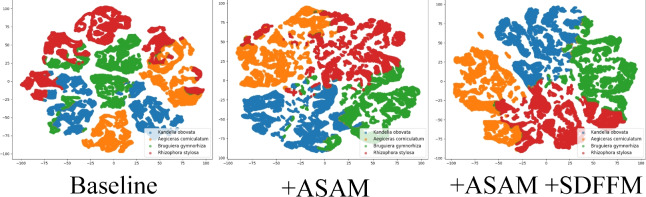
This paper also visualizes the features of mangrove species using t-SNE42, as shown in Fig. 12, comparing the effect on feature distribution after incorporating the proposed modules. The figure is divided into three sub figures, representing the feature distribution of the baseline, the addition of ASAM, and the addition of SDFFM on top of ASAM. In the baseline, the separation between different categories was small, and the same category was not clustered well, with obvious overlapping phenomena. After ASAM was added, compared with the baseline results, the overlapping phenomenon between categories was reduced, and the feature distribution became more compact and clearer. However, the boundaries were still relatively blurred, and some overlapping occurred. When ASAM and SDFFM were added to the baseline, compared with the results of only adding ASAM, the separation between different categories was further improved, and the clustering of the same category was more compact. The feature distribution was clearer, and the category boundaries were more distinct, further verifying the effectiveness of the modules proposed in this paper.
Figure 12.
t-SNE visualization of tree species features.
Potential applications
To validate the application potential of UrmsNet beyond mangrove species segmentation, we conducted additional experiments on three widely used public remote sensing datasets: LoveDA, Potsdam, and Vaihingen. These datasets represent diverse ecological environments and urban landscapes, allowing for a comprehensive test of our model’s generalization ability. The experimental results are shown in Tables 3, 4, and 5.
Table 3.
Comparative experimental results of different methods on the LoveDA dataset.
| Methods | IoU per class (%) | mIoU | ||||||
|---|---|---|---|---|---|---|---|---|
| Background | Building | Road | Water | Barren | Forest | Agriculture | (%) | |
| FCN17 | 51.31 | 62.16 | 52.41 | 58.51 | 22.86 | 39.32 | 46.61 | 47.60 |
| DeepLab v322 | 50.72 | 60.15 | 54.20 | 62.98 | 18.97 | 40.93 | 47.67 | 47.95 |
| DeepLab v3+21 | 51.48 | 62.29 | 53.57 | 56.17 | 27.23 | 40.83 | 47.52 | 48.44 |
| U-Net18 | 51.57 | 64.15 | 54.33 | 54.89 | 27.16 | 39.67 | 48.86 | 48.66 |
| U-Net++43 | 50.02 | 61.6 | 52.72 | 60.89 | 20.97 | 37.65 | 47.92 | 47.40 |
| Semantic-FPN44 | 51.36 | 63.99 | 54.37 | 56.32 | 28.25 | 41.04 | 47.79 | 49.02 |
| PSPNet45 | 52.36 | 62.4 | 48.94 | 53.65 | 22.51 | 34.68 | 47.96 | 46.07 |
| HRNet41 | 52.62 | 61.75 | 53.5 | 67.53 | 24.64 | 35.99 | 46.88 | 48.87 |
| SwinUperNet46 | 52.76 | 64.29 | 50.72 | 67.26 | 31.38 | 43.55 | 46.98 | 50.99 |
| UrmsNet | 52.54 | 62.17 | 56.44 | 70.32 | 32.15 | 44.47 | 52.97 | 53.01 |
Table 4.
Comparative experimental results of different methods on the potsdam dataset.
| Methods | IoU per class (%) | mIoU | |||||
|---|---|---|---|---|---|---|---|
| Impervious surfaces | Building | Low vegetation | Tree | Car | Clutter | (%) | |
| FCN17 | 85.19 | 90.86 | 74.12 | 78.29 | 89.53 | 31.23 | 74.87 |
| DeepLab v322 | 84.59 | 89.65 | 74.91 | 78.07 | 88.44 | 40.24 | 75.98 |
| DeepLab v3+21 | 83.76 | 89.34 | 74.62 | 77.91 | 90.21 | 34.96 | 75.13 |
| U-Net18 | 82.42 | 87.57 | 73.46 | 76.48 | 89.92 | 35.14 | 74.16 |
| U-Net++43 | 84.93 | 90.17 | 74.62 | 77.48 | 88.02 | 40.18 | 75.90 |
| Semantic-FPN44 | 82.21 | 87.41 | 73.50 | 77.91 | 89.64 | 35.42 | 74.35 |
| PSPNet45 | 83.64 | 86.90 | 71.12 | 74.13 | 87.58 | 27.83 | 71.87 |
| HRNet41 | 84.82 | 90.28 | 74.61 | 78.45 | 89.33 | 31.89 | 74.90 |
| SwinUperNet46 | 84.62 | 89.67 | 75.81 | 77.25 | 88.73 | 38.47 | 75.63 |
| UrmsNet | 86.82 | 93.67 | 76.13 | 78.64 | 91.64 | 44.14 | 78.48 |
Table 5.
Comparative experimental results of different methods on the Vaihingen dataset.
| Methods | IoU per class (%) | mIoU | ||||
|---|---|---|---|---|---|---|
| Impervious surfaces | Building | Low vegetation | Tree | Car | (%) | |
| FCN17 | 83.79 | 89.83 | 68.90 | 78.23 | 65.99 | 77.35 |
| DeepLab v322 | 83.31 | 89.91 | 68.05 | 77.92 | 65.87 | 77.01 |
| DeepLab v3+21 | 83.28 | 88.55 | 68.49 | 78.49 | 68.98 | 77.55 |
| U-Net18 | 83.33 | 88.27 | 67.87 | 77.38 | 68.59 | 77.09 |
| PSPNet45 | 83.98 | 90.76 | 68.67 | 78.52 | 72.33 | 78.85 |
| U-Net++43 | 83.53 | 88.72 | 67.40 | 77.26 | 68.17 | 77.02 |
| Semantic-FPN44 | 84.13 | 89.91 | 68.61 | 78.40 | 69.04 | 78.01 |
| HRNet41 | 80.18 | 87.95 | 67.94 | 77.89 | 73.36 | 77.44 |
| SwinUperNet46 | 81.95 | 89.34 | 68.83 | 78.34 | 72.12 | 78.12 |
| UrmsNet | 85.87 | 90.83 | 70.92 | 79.89 | 74.52 | 80.41 |
As illustrated in the tables, we compared UrmsNet with various advanced semantic segmentation methods, including FCN, DeepLab v3, DeepLab v3+, U-Net, Semantic-FPN, U-Net++, PSPNet, HRNet, and swinUperNet. Encouragingly, UrmsNet achieved the highest mIoU scores on all three datasets, demonstrating its excellent performance and strong generalization capability. Specifically, on the LoveDA dataset, UrmsNet achieved an mIoU of 53.01%, surpassing the second-best method by 2.02 percentage points. Notably, UrmsNet performed exceptionally well in categories such as roads, water bodies, barren land, forests, and agriculture. These results not only validate the model’s effectiveness but also reveal its potential application value in critical tasks such as crop classification, soil erosion monitoring, and agricultural land change detection.In terms of urban landscape analysis, UrmsNet also excelled. On the Potsdam dataset, our model achieved an mIoU of 78.48%, while on the Vaihingen dataset, it reached an impressive 80.41% mIoU, both of which are the best results among all compared methods. These outstanding performances strongly demonstrate UrmsNet’s broad application prospects in urban environment analysis, particularly in areas such as building detection, road network analysis, and urban green space planning.
These cross-dataset experimental results not only confirm UrmsNet’s excellent performance in mangrove species segmentation tasks but, more importantly, showcase the model’s robust adaptability in handling diverse geographical environments and complex urban landscapes. UrmsNet’s consistently outstanding performance across these different scenarios highlights its potential as a versatile, high-performance remote sensing image analysis tool, laying a solid foundation for future applications in multiple fields such as ecological monitoring, urban planning, and environmental protection.
Conclusion
This paper introduces a novel semantic segmentation network called UrmsNet for precise identification and segmentation of mangrove species in high-resolution drone RGB images. UrmsNet effectively addresses the challenges of morphological similarity, environmental complexity, and target size differences faced in mangrove species recognition by incorporating improved SCConv+, ASAM, and CLFFM. On the mangrove species segmentation RGB image dataset established in this study, UrmsNet achieves excellent performance with mIoU and mPA metrics of 92.21% and 95.98%, respectively, while maintaining relatively low parameter count and computational complexity. This result is comparable to the latest methods using multispectral or hyperspectral data, but significantly reduces cost and complexity. Compared to hyperspectral and multispectral approaches, UrmsNet’s RGB image-based method offers significant advantages, including lower equipment costs, simpler operation, higher data processing efficiency, smaller storage requirements, wider applicability, and higher monitoring frequency.
From a theoretical perspective, this paper makes important contributions in the following aspects: First, the design of UrmsNet breaks through the traditional limitation that high-precision mangrove species identification must rely on hyperspectral data, proving that with appropriate network structure design, RGB images can also achieve relatively accurate identification results. This finding provides new ideas for the field of remote sensing image analysis. Second, the proposal and application of ASAM demonstrate the importance of adaptive feature extraction in complex ecological environments. The ASFM within it achieves adaptive extraction and fusion of multi-scale features through three stages: decomposition, fusion, and selection. This provides a theoretical basis for solving the problems of morphological similarity and environmental complexity of mangrove species, while offering new insights into feature selection theory in ecological image analysis. The ACFM proposes a new method for adaptive fusion of multi-scale features. By learning the importance of information from different layers but at the same spatial location, it achieves more effective feature fusion, providing a new theoretical direction for solving multi-scale problems in remote sensing image analysis. CLFFM proposes a new cross-level feature fusion method by combining shallow and deep features. This method effectively utilizes detailed and boundary information from shallow features while retaining deep semantic information, providing a theoretical basis for solving scale issues and small target recognition problems common in ecological monitoring. This paper also combines advanced techniques in computer vision with specific needs of ecological monitoring, opening new directions for cross-research between ecological informatics and computer vision, promoting deep integration of these two fields. Finally, the paper proposes a new ecological monitoring method, combining periodic hyperspectral monitoring with UrmsNet-supported daily monitoring, significantly reducing cost and complexity while ensuring monitoring accuracy, providing theoretical support for large-scale ecological monitoring. These theoretical contributions not only advance mangrove monitoring technology but also provide new theoretical foundations and methodological guidance for monitoring and analysis of other complex ecosystems. At the same time, these innovative theoretical breakthroughs also provide new research directions and ideas for the development of related fields such as remote sensing image analysis, computer vision, and ecological informatics.
However, this study also has some limitations. In terms of methodology, there may be constraints on its recognition capability in certain complex environments. For instance, in conditions of extremely poor lighting, snow, rain, or severe vegetation overlap, relying solely on RGB information might make it difficult to accurately distinguish different mangrove species, still necessitating the involvement of multispectral and hyperspectral data. Furthermore, the current method is primarily optimized for static images and may require further improvements for monitoring dynamic changes (such as the impact of tidal changes on mangrove morphology). Regarding data, although a new dataset was constructed, the geographical distribution and seasonal variation of samples may not be comprehensive enough, potentially affecting the model’s generalization ability under different environmental conditions. Additionally, while UrmsNet performs well on this dataset and the LoveDA, Potsdam, and Vaihingen datasets, its application effectiveness in non-mangrove ecosystems still requires in-depth study, especially in environments with significant differences in vegetation density and species diversity.
To fully leverage UrmsNet’s application value in practical ecological monitoring, our future work will focus on several key directions. Firstly, we will develop a comprehensive, flexible, and extensible application programming interface (API) that allows UrmsNet to be easily integrated into existing ecological monitoring workflows and connect with satellite or drone data collection systems, thereby improving monitoring efficiency and response speed. Simultaneously, we will emphasize the standardized processing of output results, ensuring that analysis results conform to industry-wide standards and can be directly imported into mainstream geographic information system (GIS) software and professional ecological data management platforms, thus expanding the scope of application. Additionally, future research will explore UrmsNet’s potential applications in monitoring other ecosystems, including its combined use with hyperspectral data and integration with other remote sensing technologies and ecological methods. We also plan to explore the possibility of fusing UrmsNet with other sensor data, develop versions for time-series data, expand the dataset to cover more geographical regions, seasonal variations, and extreme weather conditions, and investigate the method’s application in broader ecosystem monitoring. Finally, we will actively explore unsupervised mangrove species segmentation methods. These efforts will help further enhance UrmsNet’s robustness and versatility, making greater contributions to the field of ecological monitoring.
In summary, this paper presents an innovative solution for mangrove ecosystem monitoring that is efficient, accurate, and economical. It not only provides robust technical support for the protection and sustainable management of mangrove forests but also contributes valuable data resources for related research. UrmsNet pioneers a new technological approach for large-scale, low-cost monitoring of important ecosystems such as mangrove forests, while also offering valuable insights for research and conservation efforts in other similar ecosystems. Although some limitations currently exist, through continuous optimization and functional expansion, UrmsNet has the potential to become a key tool in the field of ecological monitoring and environmental protection, demonstrating the significant role of technological innovation in promoting ecological sustainable development.
Acknowledgements
This work was supported by Guangxi Science and Technology Major Project(AA19254016) and Beihai Science and Technology Bureau Project: Bei Kehe 2023158004 Project support.
Author contributions
Xin Wang and Yu Zhang and Yi Feng proposed the network architecture design and framework. Yu Zhang and Jingye Cai performed the experiments and analyzed the data. Yu Zhang and Qin Qin wrote the manuscript.Jingke Yan revised the paper and provided valuable advice regarding the experiments. All authors have read and approved the fnal manuscript.
Data availability
Readers can access our data by sending an email to the corresponding author.
Code availability
Readers can access our code by sending an email to the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abidin, Z., Setiawan, B., Muhaimin, A. W. & Shinta, A. The role of coastal biodiversity conservation on sustainability and environmental awareness in mangrove ecosystem of southern malang, indonesia. Biodiversitas J. Biol. Divers.22 (2021).
- 2.Lee, S. Y. et al. Ecological role and services of tropical mangrove ecosystems: a reassessment. Glob. Ecol. Biogeogr.23, 726–743 (2014). [Google Scholar]
- 3.Kuenzer, C., Bluemel, A., Gebhardt, S., Quoc, T. V. & Dech, S. Remote sensing of mangrove ecosystems: A review. Remote Sens.3, 878–928 (2011). [Google Scholar]
- 4.Schaepdryver, K. H., Goossens, W., Naseef, A., Kalpuzha Ashtamoorthy, S. & Steppe, K. Foliar water uptake capacity in six mangrove species. Forests13, 951 (2022). [Google Scholar]
- 5.Kathiresan, K. Mangroves: types and importance. Mangroves Ecol. Biodivers. Manag. 1–31 (2021).
- 6.Menezes, M., Berger, U. & Worbes, M. Annual growth rings and long-term growth patterns of mangrove trees from the bragança peninsula, north brazil. Wetlands Ecol. Manage.11, 233–242 (2003). [Google Scholar]
- 7.Sadono, R., Soeprijadi, D., Susanti, A., Wirabuana, P. Y. A. P. & Matatula, J. Species composition and growth performance of mangrove forest at the coast of tanah merah, east nusa tenggara. Biodiversitas Journal of Biological Diversity21 (2020).
- 8.Krauss, K. W. & Osland, M. J. Tropical cyclones and the organization of mangrove forests: a review. Ann. Bot.125, 213–234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treviño, M. “the mangrove is like a friend’’: Local perspectives of mangrove cultural ecosystem services among mangrove users in northern ecuador. Hum. Ecol.50, 863–878 (2022). [Google Scholar]
- 10.Zimudzi, E., Sanders, I., Rollings, N. & Omlin, C. W. Remote sensing of mangroves using unmanned aerial vehicles: Current state and future directions. J. Spat. Sci.66, 195–212 (2021). [Google Scholar]
- 11.Adade, R., Aibinu, A. M., Ekumah, B. & Asaana, J. Unmanned aerial vehicle (uav) applications in coastal zone management-a review. Environ. Monit. Assess.193, 1–12 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Wang, H., Liu, Z., Hu, G., Wang, X. & Han, Z. Offline meta-reinforcement learning for active pantograph control in high-speed railways. IEEE Trans. Ind. Inf. (2024). [DOI] [PubMed]
- 13.Yan, J. et al. Transformer and graph convolution-based unsupervised detection of machine anomalous sound under domain shifts. IEEE Trans. Emerg. Topics Comput. Intell. (2024).
- 14.Bhatnagar, S., Gill, L. & Ghosh, B. Drone image segmentation using machine and deep learning for mapping raised bog vegetation communities. Remote Sens.12, 2602 (2020). [Google Scholar]
- 15.Song, Z., Zhang, Z., Yang, S., Ding, D. & Ning, J. Identifying sunflower lodging based on image fusion and deep semantic segmentation with uav remote sensing imaging. Comput. Electron. Agric.179, 105812 (2020). [Google Scholar]
- 16.Li, G. et al. Extraction of sunflower lodging information based on uav multi-spectral remote sensing and deep learning. Remote Sens.13, 2721 (2021). [Google Scholar]
- 17.Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, 3431–3440 (2015). [DOI] [PubMed]
- 18.Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18, 234–241 (Springer, 2015).
- 19.Zhou, Z., Chen, B., Xu, R. & Fang, W. Mangrove species identification based on uav hyperspectral features. Acta Oceanol. Sin.43, 137–145 (2021). [Google Scholar]
- 20.Gao, C. et al. Mangrove species identification using worldview-2 satellite imagery. Wetland Sci.21, 627 (2023). [Google Scholar]
- 21.Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV), 801–818 (2018).
- 22.Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K. & Yuille, A. L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell.40, 834–848 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Xiao, T., Liu, Y., Zhou, B., Jiang, Y. & Sun, J. Unified perceptual parsing for scene understanding. In Proceedings of the European conference on computer vision (ECCV), 418–434 (2018).
- 24.Zhao, H., Shi, J., Qi, X., Wang, X. & Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2881–2890 (2017).
- 25.Lin, T.-Y. et al. Feature pyramid networks for object detection. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2117–2125 (2017).
- 26.Valderrama-Landeros, L., Flores-de Santiago, F., Kovacs, J. & Flores-Verdugo, F. An assessment of commonly employed satellite-based remote sensors for mapping mangrove species in mexico using an ndvi-based classification scheme. Environ. Monit. Assess.190, 1–13 (2018). [DOI] [PubMed]
- 27.Peng, L. et al. Combining gf-2 and rapideye satellite data for mapping mangrove species using ensemble machine-learning methods. Int. J. Remote Sens.41, 813–838 (2020). [Google Scholar]
- 28.Rahmandhana, A. D., Kamal, M. & Wicaksono, P. Spectral reflectance-based mangrove species mapping from worldview-2 imagery of karimunjawa and kemujan island, central java province, indonesia. Remote Sens.14, 183 (2022). [Google Scholar]
- 29.Yang, Y. et al. Fine-scale mangrove species classification based on uav multispectral and hyperspectral remote sensing using machine learning. Remote Sens.16, 3093 (2024). [Google Scholar]
- 30.Zhen, J. et al. Performance of xgboost ensemble learning algorithm for mangrove species classification with multisource spaceborne remote sensing data. J. Remote Sens.4, 0146 (2024). [Google Scholar]
- 31.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778 (2016).
- 32.Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint[SPACE]arXiv:1409.1556 (2014).
- 33.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2818–2826 (2016).
- 34.Li, J., Wen, Y. & He, L. Scconv: spatial and channel reconstruction convolution for feature redundancy. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 6153–6162 (2023).
- 35.Li, Y. et al. Large selective kernel network for remote sensing object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 16794–16805 (2023).
- 36.Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst.25 (2012).
- 37.Howard, A. G. et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint[SPACE]arXiv:1704.04861 (2017).
- 38.Russell, B. C., Torralba, A., Murphy, K. P. & Freeman, W. T. Labelme: a database and web-based tool for image annotation. Int. J. Comput. Vision77, 157–173 (2008). [Google Scholar]
- 39.Liu, Z. et al. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF international conference on computer vision, 10012–10022 (2021).
- 40.Liu, Z. et al. A convnet for the 2020s. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 11976–11986 (2022).
- 41.Sun, K., Xiao, B., Liu, D. & Wang, J. Deep high-resolution representation learning for human pose estimation. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 5693–5703 (2019).
- 42.Van der Maaten, L. & Hinton, G. Visualizing data using t-sne. J. Mach. Learn. Res.9 (2008).
- 43.Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N. & Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4, 3–11 (Springer, 2018). [DOI] [PMC free article] [PubMed]
- 44.Kirillov, A., Girshick, R., He, K. & Dollar, P. Panoptic feature pyramid networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2019).
- 45.Zhao, H., Shi, J., Qi, X., Wang, X. & Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2881–2890 (2017).
- 46.Wang, Z., Li, J., Tan, Z., Liu, X. & Li, M. Swin-upernet: A semantic segmentation model for mangroves and spartina alterniflora loisel based on upernet. Electronics12, 1111 (2023). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Readers can access our data by sending an email to the corresponding author.
Readers can access our code by sending an email to the corresponding author.